Basic Information
-
Targeting strategy

-
Gene targeting strategy for B-hIFNGR1/hIFNG/hIFNGR2 mice.
- The full coding sequence of mouse Ifng gene was replaced by counterpart of human IFNG gene in B-hIFNGR1/hIFNG/hIFNGR2 mice.
- The sequence that encoding the extracellular region of mouse Ifngr1 gene was replaced by counterpart human IFNGR1 gene in B-hIFNGR1/hIFNG/hIFNGR2 mice.
- The sequence that encoding the extracellular region of mouse Ifngr2 gene was replaced by counterpart human IFNGR2 gene in B-hIFNGR1/hIFNG/hIFNGR2 mice.
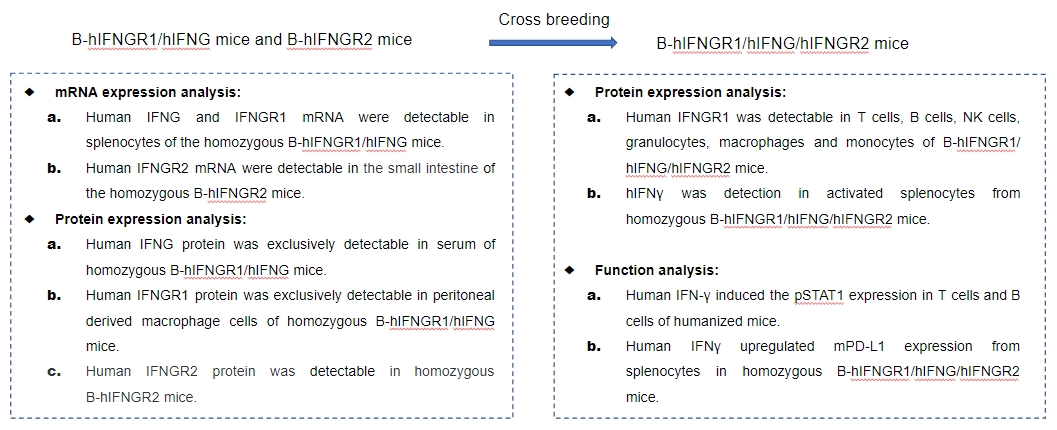
Note: B-hIFNGR1/hIFNG/hIFNGR2 mice was obtained by cross breeding B-hIFNGR1/hIFNG mice with B-hIFNGR2 mice.
-
mRNA expression analysis

-

IFNG and IFNGR1 expression analysis in B-hIFNGR1/hIFNG mice by RT-PCR. Human IFNG and IFNGR1 mRNA were detectable in splenocytes of the homozygous B-hIFNGR1/hIFNG mice (H/H) but not in wild-type mice (+/+).
-
Protein expression analysis

-
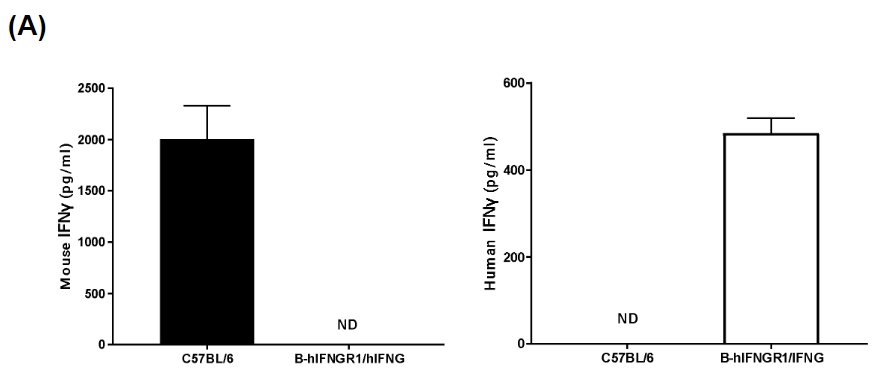
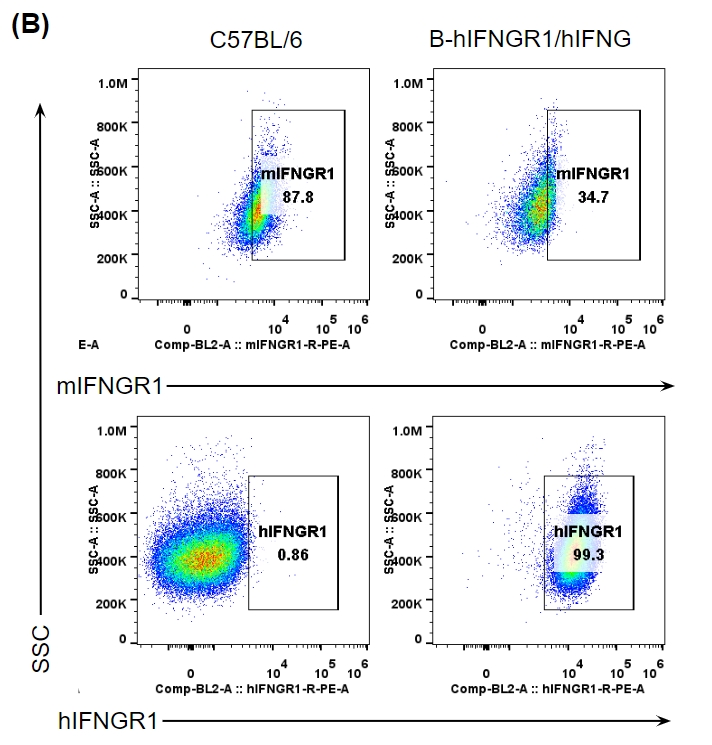
IFNG and IFNGR1 expression analysis in B-hIFNGR1/IFNG mice. (A) IFNG expression. Serum was collected from wild-type C57BL/6 mice and homozygous B-hIFNGR1/hIFNG mice (anti-CD3ε antibody, 2 hours, in vivo) and analyzed by ELISA with species-specific IFNG ELISA kit. Human IFNG was exclusively detectable in homozygous B-hIFNGR1/hIFNG mice but not in wild-type mice. (B) IFNGR1 expression. Peritoneal derived macrophage cells were collected, and analyzed by flow cytometry with anti-IFNGR1 antibody. Human IFNGR1 was detectable in B-hIFNGR1/hIFNG mice.
-
mRNA and protein expression analysis

-
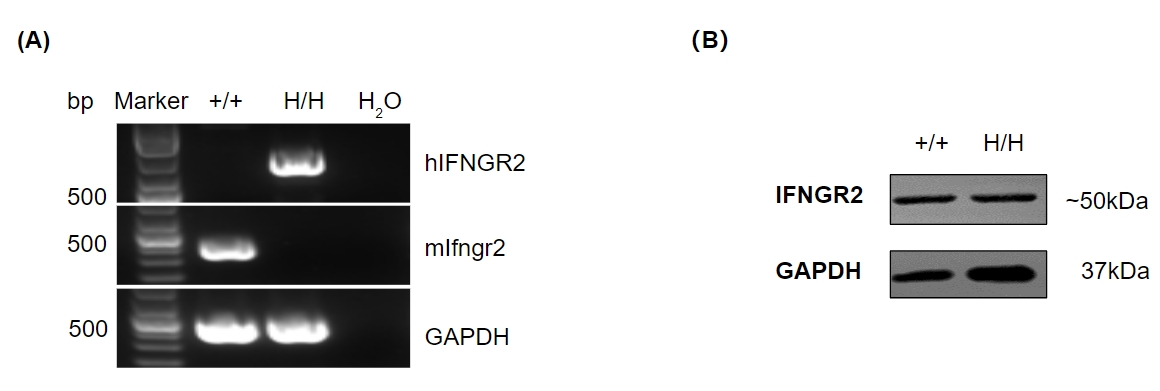
Strain specific analysis of IFNGR2 gene expression in wild-type mice and B-hIFNGR2 mice by RT-PCR and western blotting. (A) mRNA expression. Mouse Ifngr2 mRNA was exclusively detectable in the small intestine of wild-type mice (+/+). Human IFNGR2 mRNA was exclusively detectable in homozygous B-hIFNGR2 mice (H/H). (B) Protein expression. Skeletal muscle was collected from wild-type C57BL/6 mice (+/+) and homozygous B-hIFNGR2 mice (H/H), and analyzed by western blotting with anti-IFNGR2 antibody which can recognize both human and mouse IFNGR2. The results show that IFNGR2 protein can be detected in homozygous B-hIFNGR2 mice.
-
Activated splenocytes from triple-KI mice produce human IFNγ

-
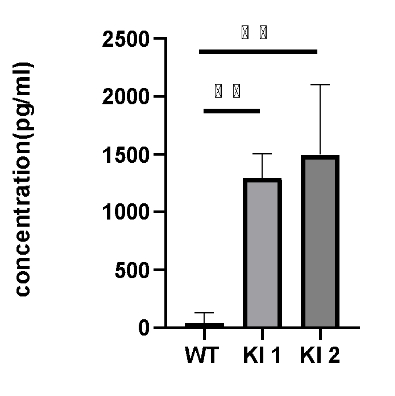
Analysis of hIFNγ production in activated splenocytes from homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice (Triple-KI). Cells were seeded in a 96-wells plate for 0.5X106 cells/well. The plate was precoated with 8μg/mL anti-mCD3 mAb at 4 ℃ for overnight. Then incubated the cells at 37℃ 5% CO2 for 5 days. Anti-CD28 at 1μg/mL and 100U hIL2 were added 16 hours after culture. Get the supernatant for ELISA analysis. hIFNγ was detection in triple-KI mice but not in wild-type mice.
*The experiment is verified by the cooperation partner.
-
IFNGR1 expression in different immune cells

-
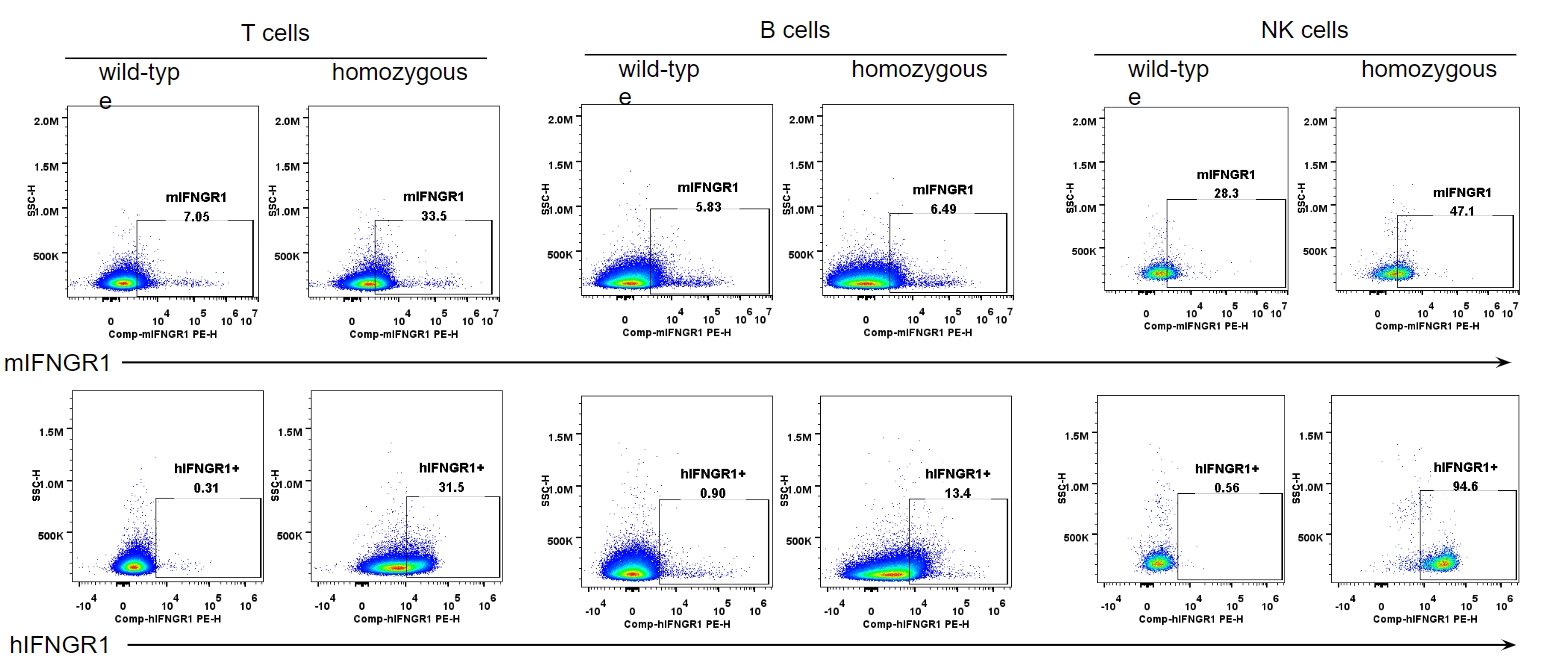
IFNGR1 expression analysis in B-hIFNGR1/IFNG/hIFNGR2 mice. Splenocytes were collected from wild-type C57BL/6 mice and homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice, and analyzed by flow cytometry with anti-IFNGR1 antibody. Human IFNGR1 was detectable in T cells, B cells and NK cells of B-hIFNGR1/hIFNG/hIFNGR2 mice.
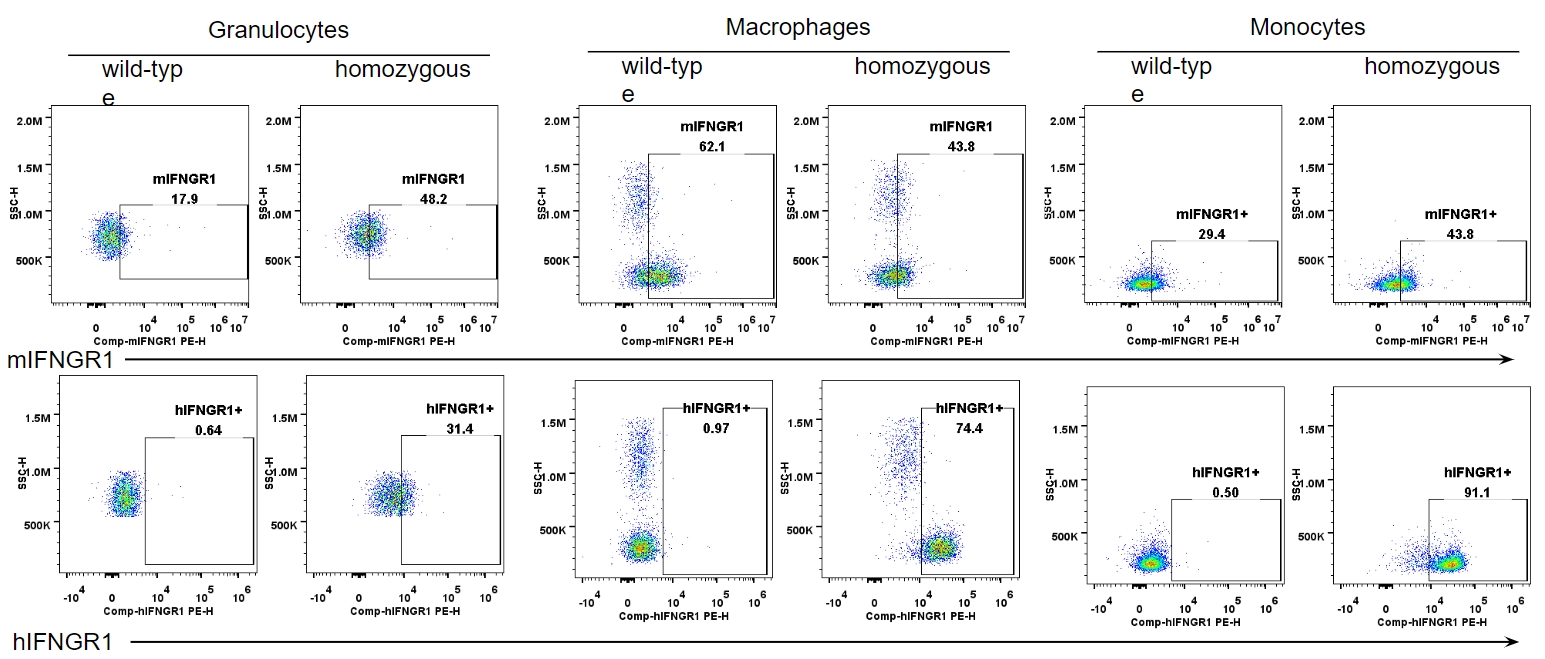
IFNGR1 expression analysis in B-hIFNGR1/hIFNG/hIFNGR2 mice. Splenocytes were collected from wild-type C57BL/6 mice and homozygous B-hIFNGR1/IFNG/hIFNGR2 mice, and analyzed by flow cytometry with anti-IFNGR1 antibody. Human IFNGR1 was detectable in granulocytes, macrophages and monocytes of B-hIFNGR1/hIFNG/hIFNGR2 mice.
-
IFNγ induced phosphorylation of STAT1 in T, B and NK cells

-
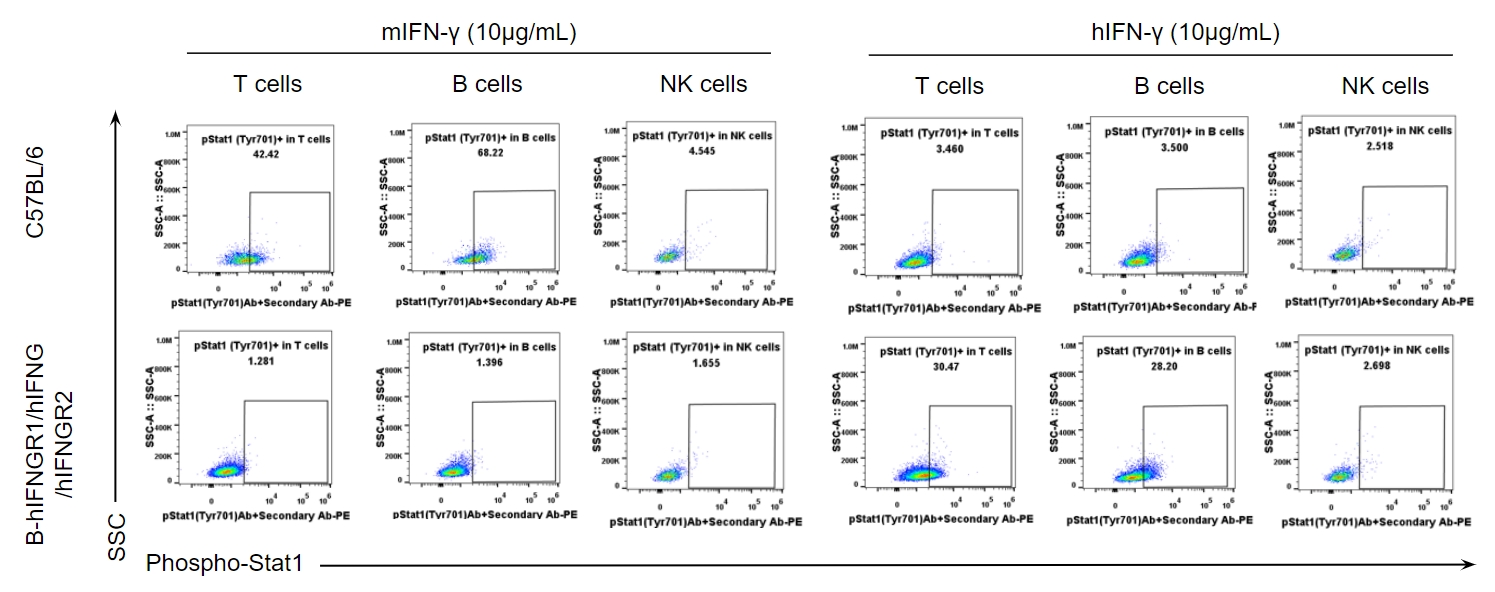
Analysis of the IFNγ induced pSTAT1(Tyr701) in wild-type C57BL/6 mice and homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice by flow cytometry. Splenocytes from wild-type mice and homozygous mice (female, 7-week-old) were treated with IFNγ for ex vivo, and then flow cytometry was performed to analyze the phosphorylation of STAT1. Mouse IFN-γ induced the expression of pSTAT1 in T and B cells of wild-type mice but not humanized mice. Human IFN-γ induced the expression of pSTAT1 in T and B cells of humanized mice but not wild-type mice. Among NK cells, IFN-γ not induced the STAT1 phosphorylation in both mice.
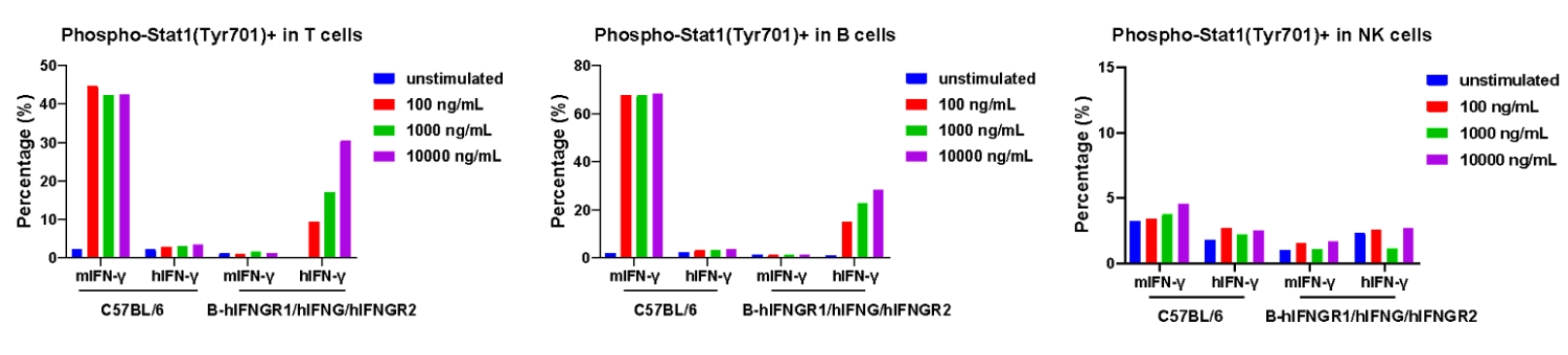
Analysis of the IFNγ induced pSTAT1(Tyr701) in wild-type C57BL/6 mice and homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice by flow cytometry. Splenocytes from wild-type mice and homozygous mice (female, 7-week-old) were treated with IFNγ in doses shown in the panel for ex vivo, and then flow cytometry was performed to analyze the phosphorylation of STAT1. Mouse IFNγ induced the expression of pSTAT1 in T and B cells of wild-type mice but not humanized mice. Human IFNγ induced the expression of pSTAT1 in T and B cells of humanized mice but not wild-type mice. Among NK cells, IFNγ not induced the STAT1 phosphorylation in both mice.
-
hIFNγ upregulates mPD-L1 in splenocytes from triple-KI mice, not WT mice

-
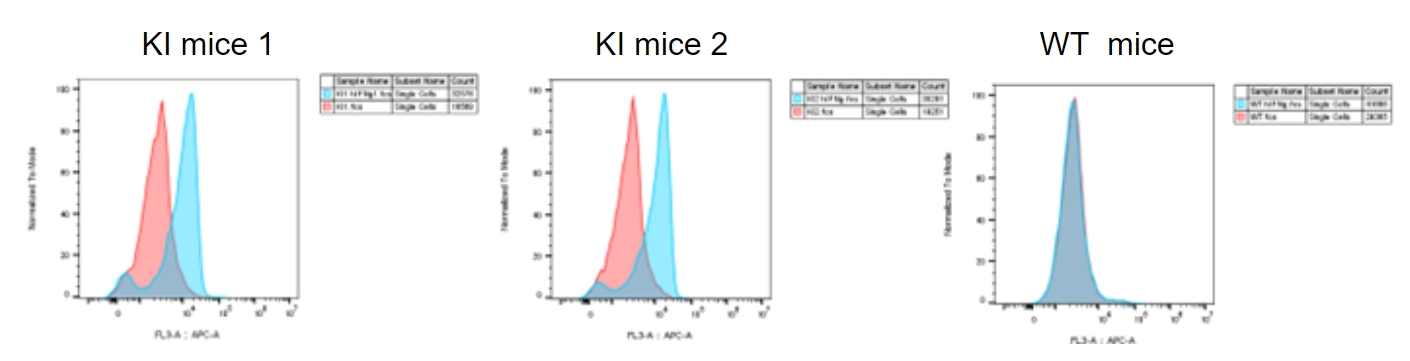
hIFNγ upregulated mPD-L1 in splenocytes from homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice (Triple-KI). Splenocytes at 0.5×106/well (96-well plate) were treated with 100ng/mL hIFNγ for 24 hours at 37℃. Cells were collected and assessed for mPD-L1 expression by flow cytometry. As shown, hIFNγ induced the expression of mPD-L1 in triple-KI mice but not in wild-type mice.
*The experiment is verified by the cooperation partner.
-
Analysis of leukocytes cell subpopulation in spleen

-

Analysis of spleen leukocyte subpopulations by flow cytometry. Splenocytes were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in spleen

-
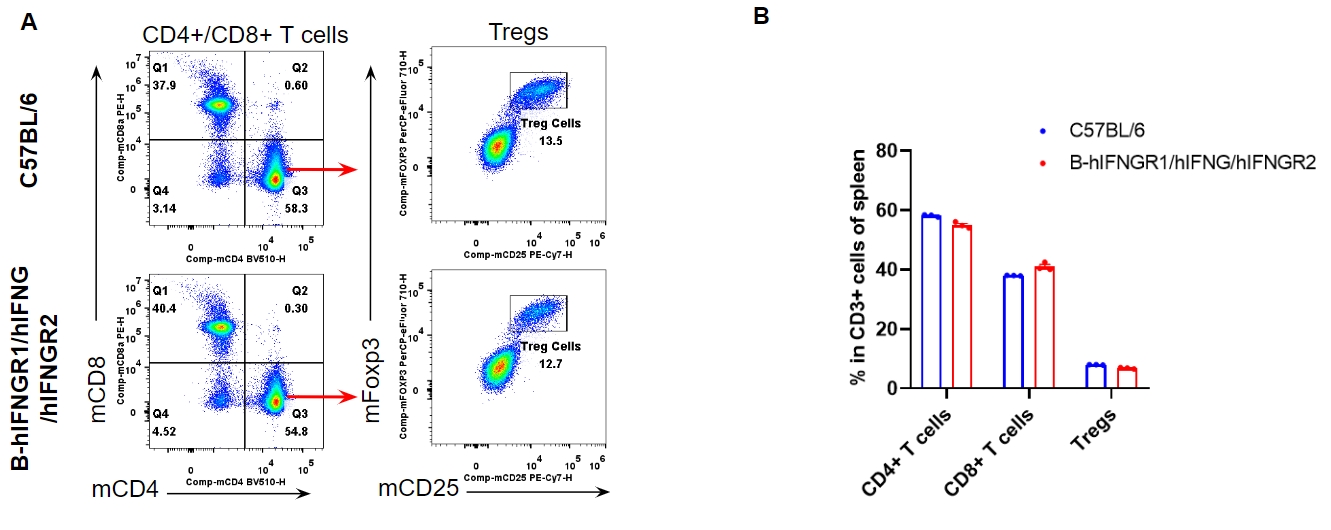
Analysis of spleen T cell subpopulations by flow cytometry. Splenocytes were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in blood

-
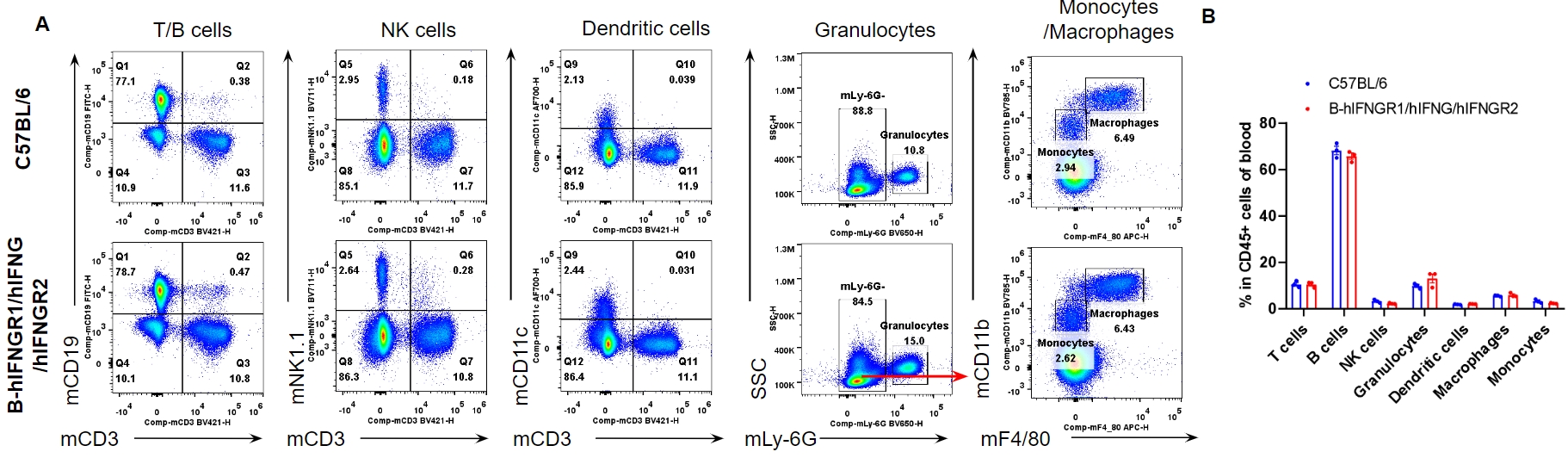
Analysis of blood leukocyte subpopulations by flow cytometry. Blood cells were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the blood was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in blood

-

Analysis of blood T cell subpopulations by flow cytometry. Blood cells were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the blood was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in lymph node

-
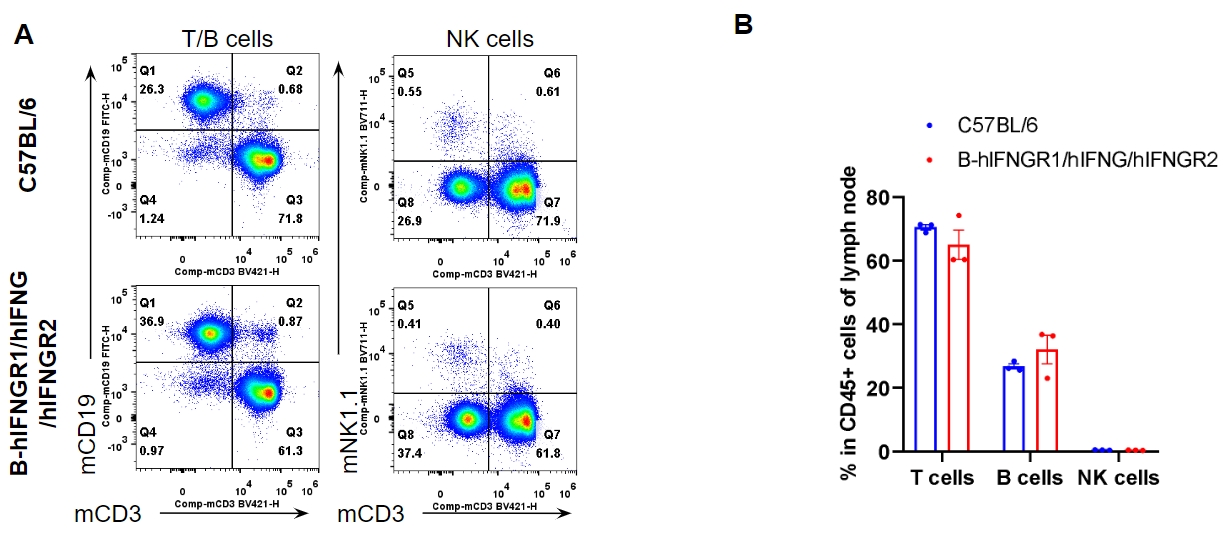
Analysis of lymph node leukocyte subpopulations by flow cytometry. Lymph node were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the lymph node was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells and NK cells in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in lymph node

-

Analysis of lymph node T cell subpopulations by flow cytometry. Lymph node were isolated from female C57BL/6 and B-hIFNGR1/hIFNG/hIFNGR2 mice (n=3, 14-week-old). Flow cytometry analysis of the lymph node was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hIFNGR1/hIFNG/hIFNGR2 mice were similar to those in the C57BL/6 mice. Values are expressed as mean ± SEM.
-
Blood biochemical detection

-
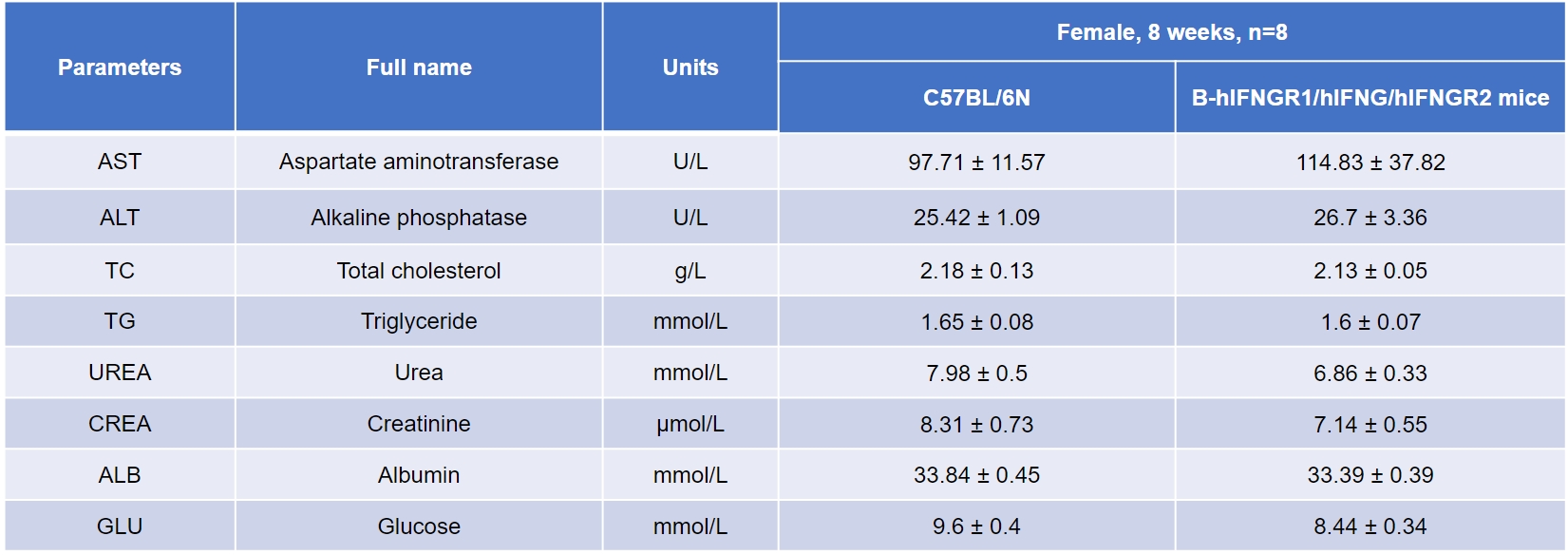
-
Blood routine detection

-
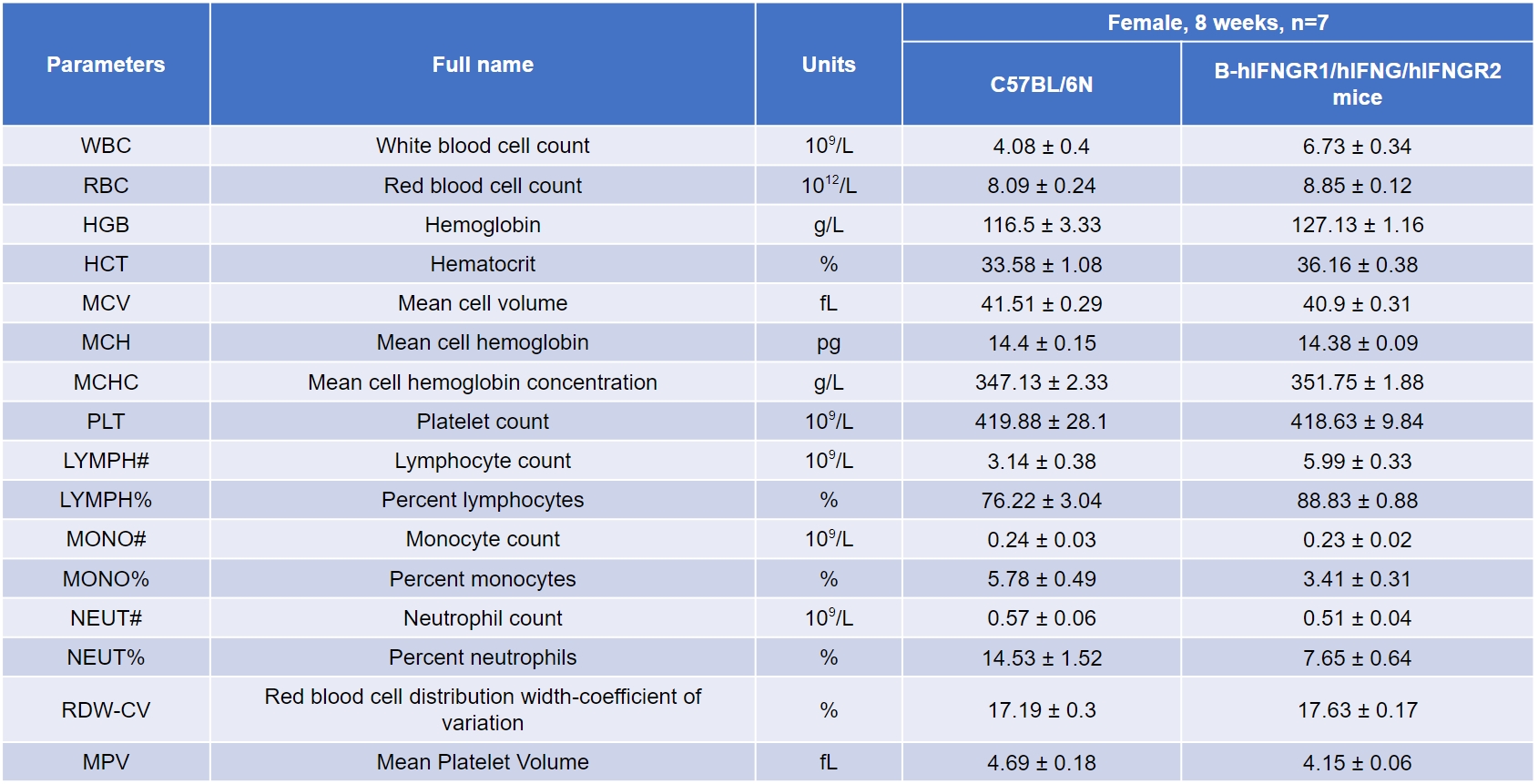
-
Summary

-