Basic Information
-
Details

-
Protein expression analysis

Strain specific TNFα expression analysis in homozygous B-hTNFA/hIL17A mice by ELISA.
Serum were collected from WT and homozygous B-hTNFA/hIL17A (H/H) mice stimulated with LPS in vivo, and analyzed by ELISA with species-specific TNFα ELISA kit. Mouse TNFα was detectable in WT mice. Human TNFα was exclusively detectable in homozygous B-hTNFA/hIL17A but not WT mice.
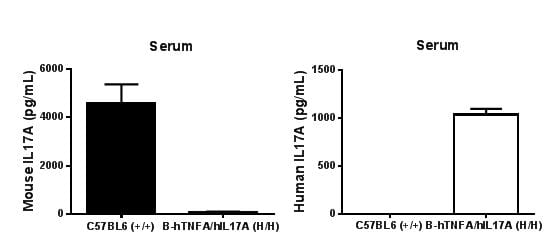
Strain specific IL17A expression analysis in homozygous B-hTNFA/hIL17A mice by ELISA.
Serum were collected from WT and homozygous B-hTNFA/hIL17A (H/H) mice stimulated with LPS in vivo, and analyzed by ELISA with species-specific IL17A ELISA kit. Mouse IL17A was detectable in WT mice. Human IL17A was exclusively detectable in homozygous B-hTNFA/hIL17A but not WT mice.
Phenotypic analysis
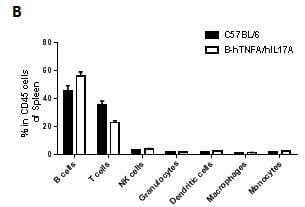
Analysis of spleen leukocytes cell subpopulations in B-hTNFA/hIL17A mice
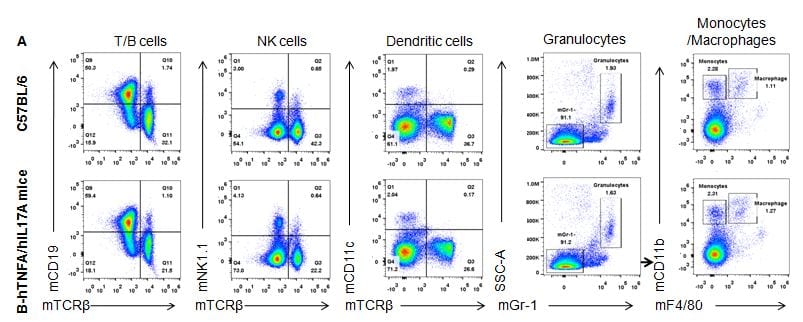
Analysis of spleen T cell subpopulations in B-hTNFA/hIL17A mice
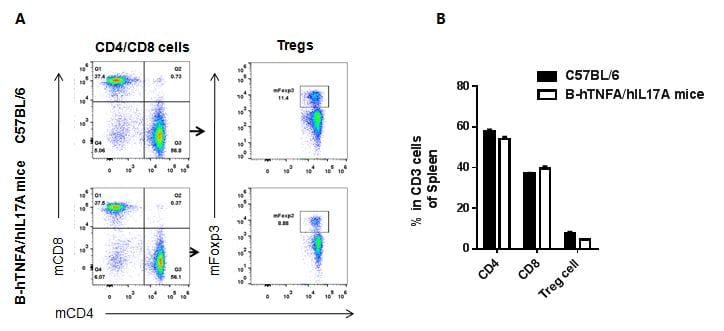
Analysis of spleen T cell subpopulations by FACS
Splenocytes were isolated from female C57BL/6 and B-hTNFA/hIL17A mice (n=3, 6 week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3 T cell population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of CD8, CD4, and Treg cells in homozygous B-hTNFA/hIL17A mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hTNFA/hIL17A in place of its mouse counterpart does not change the overall development, differentiation or distribution of these T cell sub types in spleen. Values are expressed as mean ± SEM.
Analysis of lymph node leukocytes cell subpopulations in B-hTNFA/hIL17A mice
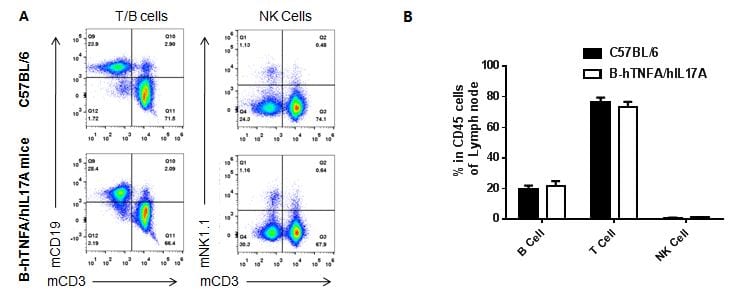
Analysis of lymph node leukocyte subpopulations by FACS
Leukocytes were isolated from female C57BL/6 and B-hTNFA/hIL17A mice (n=3, 6 week-old) Flow cytometry analysis of the leukocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for CD45 population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T, B and NK cells in homozygous B-hTNFA/hIL17A mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hTNFA/hIL17A in place of its mouse counterpart does not change the overall development, differentiation or distribution of these cell types in lymph node.
Analysis of lymph node T cell subpopulations in B-hTNFA/hIL17A mice
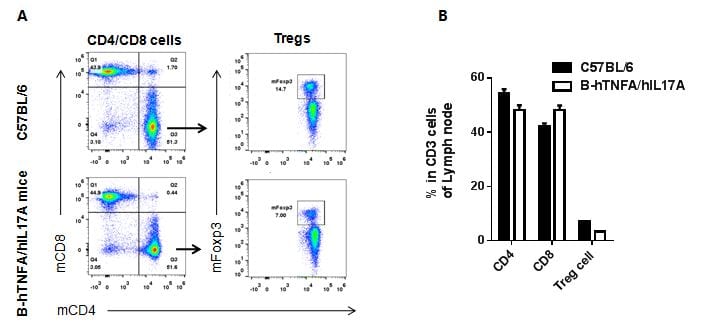
Analysis of lymph node T cell subpopulations by FACS
Leukocytes were isolated from female C57BL/6 and B-hTNFA/hIL17A mice (n=3, 6 week-old). Flow cytometry analysis of the leukocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3 T cell population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of CD8, CD4, and Treg cells in homozygous B-hTNFA/hIL17A mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hTNFA/hIL17A in place of its mouse counterpart does not change the overall development, differentiation or distribution of these T cell sub types in lymph node. Values are expressed as mean ± SEM.


