Basic Information
Description
The exogenous promoter and chimeric CDS composed of mouse signal peptide, human extracellular region, moue transmembrane region and mouse intracellular region of LAIR1 was inserted to replace part of mouse exon 3 and intron 3. Human LAIR1 is highly expressed on the surface of B-hLAIR1-luc EL4 cells.
-
Targeting Strategy

-
Gene targeting strategy for B-hLAIR1-luc EL4 cells. The exogenous promoter and chimeric CDS composed of mouse signal peptide, human extracellular region, moue transmembrane region and mouse intracellular region of LAIR1 was inserted to replace part of mouse exon 3 and intron 3. The insertion disrupts the endogenous murine Lair1 gene, resulting in a non-functional transcript.
-
Protein Expression Analysis

-
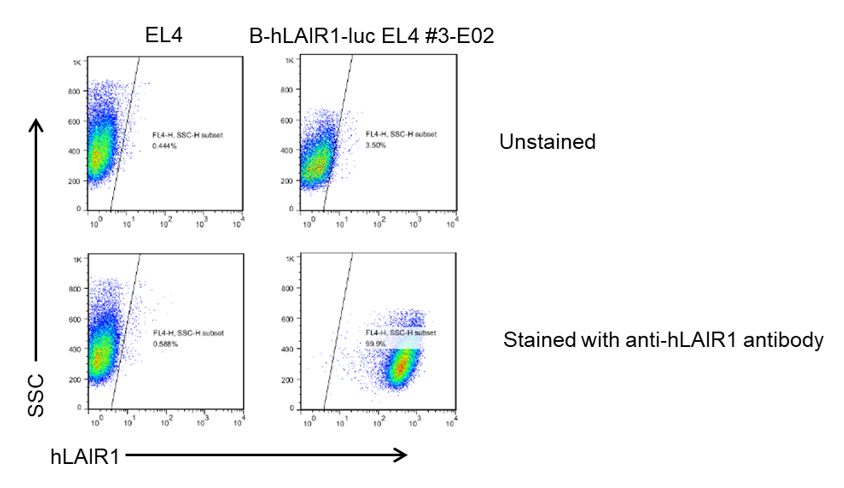
LAIR1 expression analysis in B-hLAIR1-luc EL4 cells by flow cytometry. Single cell suspensions from wild-type EL4 and B-hLAIR1-luc EL4 cultures were stained with species-specific anti-LAIR1 antibody. Human LAIR1 was detected on the surface of B-hLAIR1-luc EL4 cells but not wild-type EL4 cells. The 3-E02 clone of B-hLAIR1-luc EL4 cells was used for in vivo experiments.
-
Tumor growth curve & Body weight changes

-
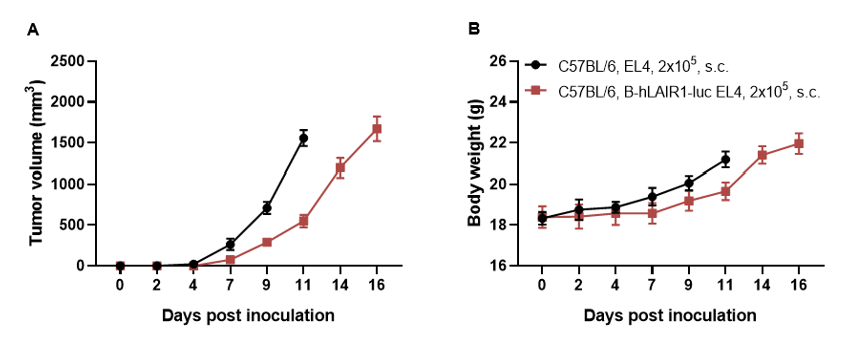
Subcutaneous homograft tumor growth of B-hLAIR1-luc EL4 cells. B-hLAIR1-luc EL4 cells (2×105) and wild-type EL4 cells (2×105) were subcutaneously implanted into C57BL/6 mice (female, 7-week-old, n=5). Tumor volume and body weight were measured twice a week. (A) Average tumor volume. (B) Body weight. Volume was expressed in mm3 using the formula: V=0.5 X long diameter X short diameter2. As shown in panel A, B-hLAIR1-luc EL4 cells were able to establish tumors in vivo and can be used for efficacy studies. Values are expressed as mean ± SEM.
-
Protein expression analysis of tumor cells

-
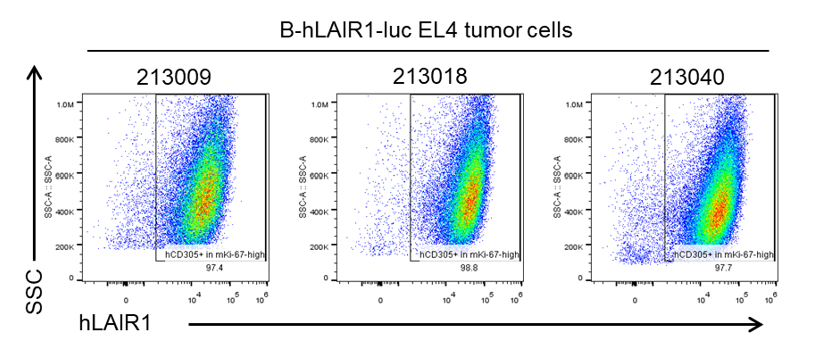
B-hLAIR1-luc EL4 cells were subcutaneously transplanted into C57BL/6 mice (n=5). At the end of the experiment, tumor cells were harvested and assessed for human LAIR1 expression by flow cytometry. As shown, human LAIR1 was highly expressed on the surface of tumor cells. Therefore, B-hLAIR1-luc EL4 cells can be used for in vivo efficacy studies of novel LAIR1 therapeutics.


