Basic Information
-
Targeting strategy

-
Gene targeting strategy for B-hCD16A/hNKP46 mice. The regulatory region and exons 1~5 of mouse Fcgr4 gene that encode the full-length protein were replaced by human FCGR3A(F158V) exons 1~6 and regulatory region in B-hCD16A/hNKP46 mice. The exons 1~7 of mouse NKp46 gene that encode the extracellular domain were replaced by human NKP46 exons 1~7 in B-hCD16A/hNKP46 mice.
-
Protein expression analysis of NKP46

-
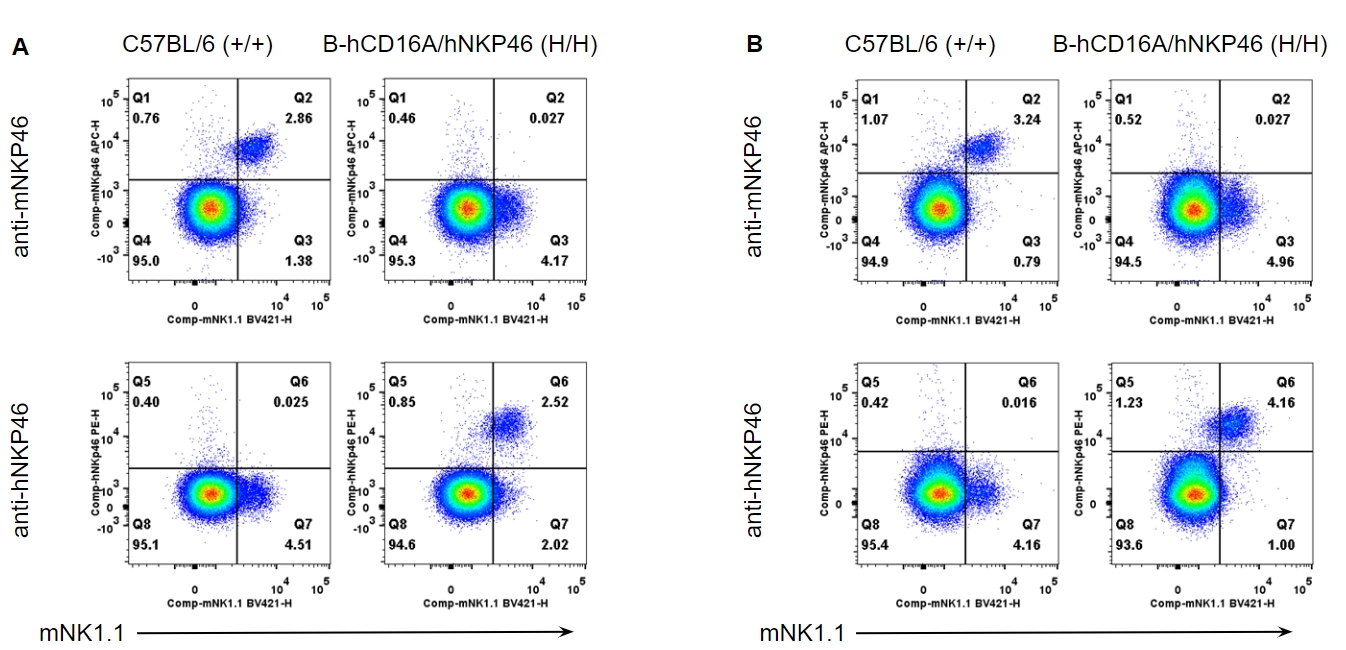
Strain specific NKP46 expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific NKP46 antibody. Mouse NKP46 was detectable in wild-type mice. Human NKP46 was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-T cells

-
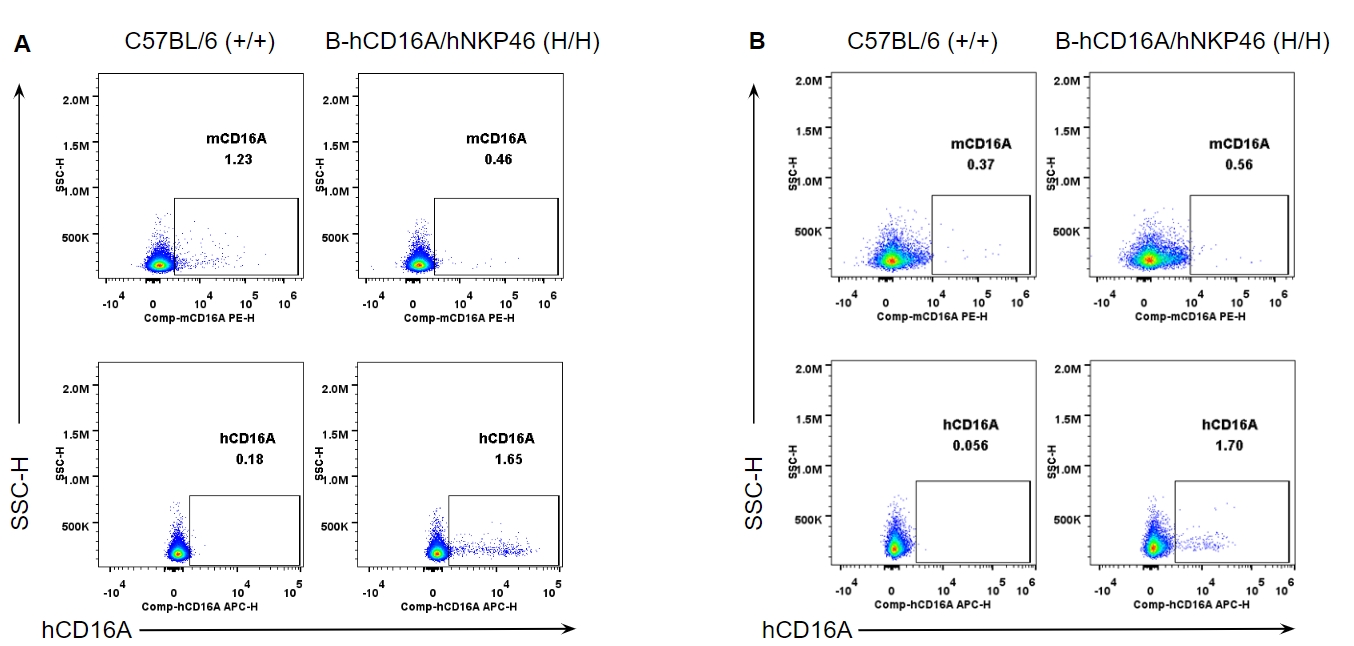
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was not detectable in wild-type mice. Human CD16A was not detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-B cells

-
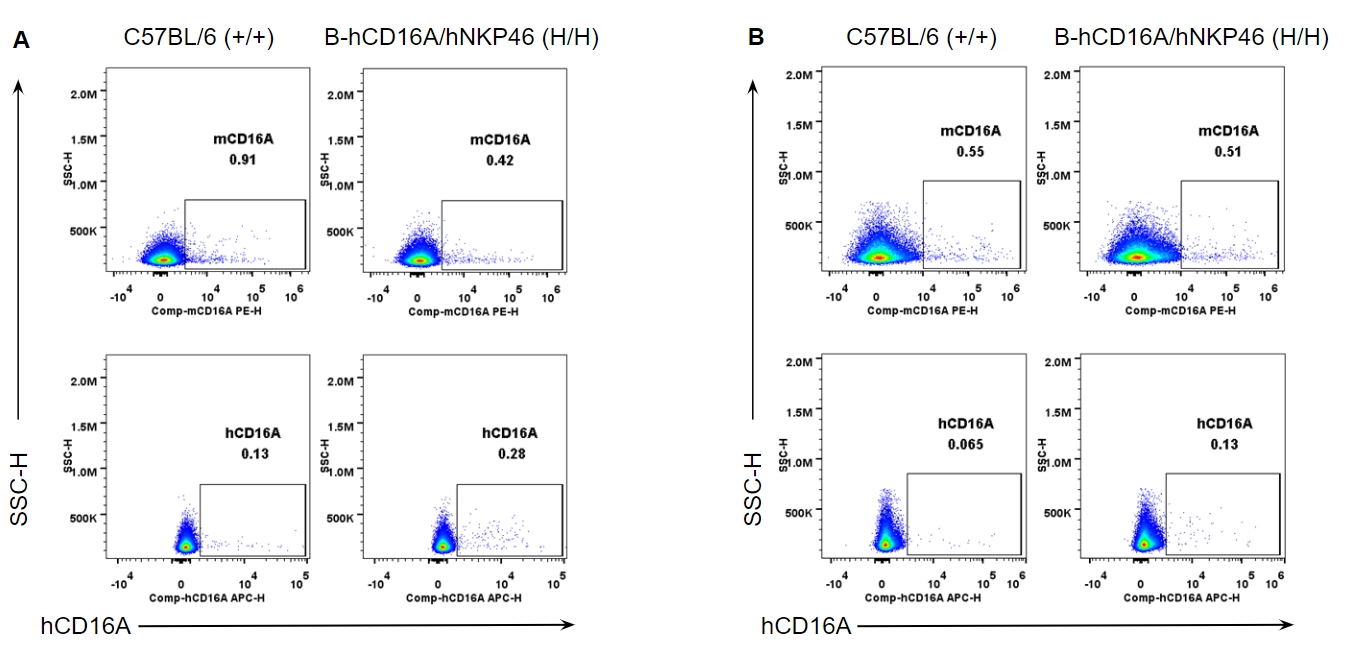
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was not detectable in wild-type mice. Human CD16A was not detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-NK cells

-
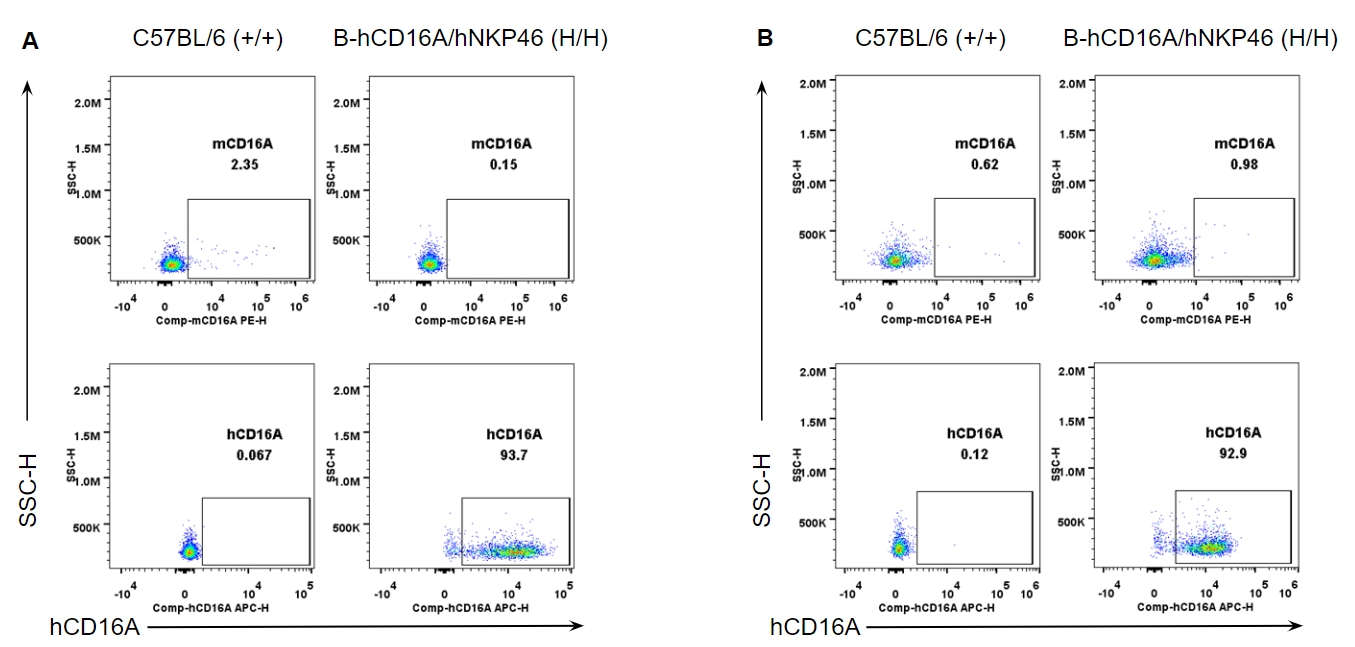
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was not detectable in wild-type mice. Human CD16A was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-DC cells

-
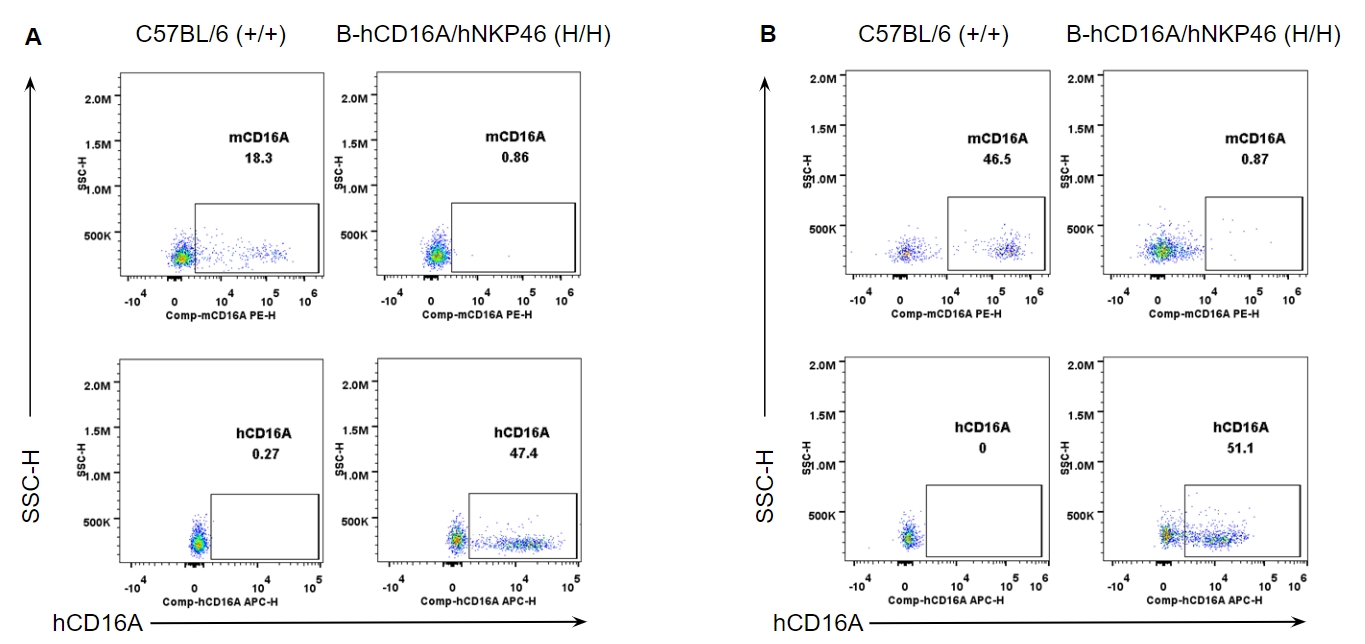
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was detectable in wild-type mice. Human CD16A was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-monocytes

-
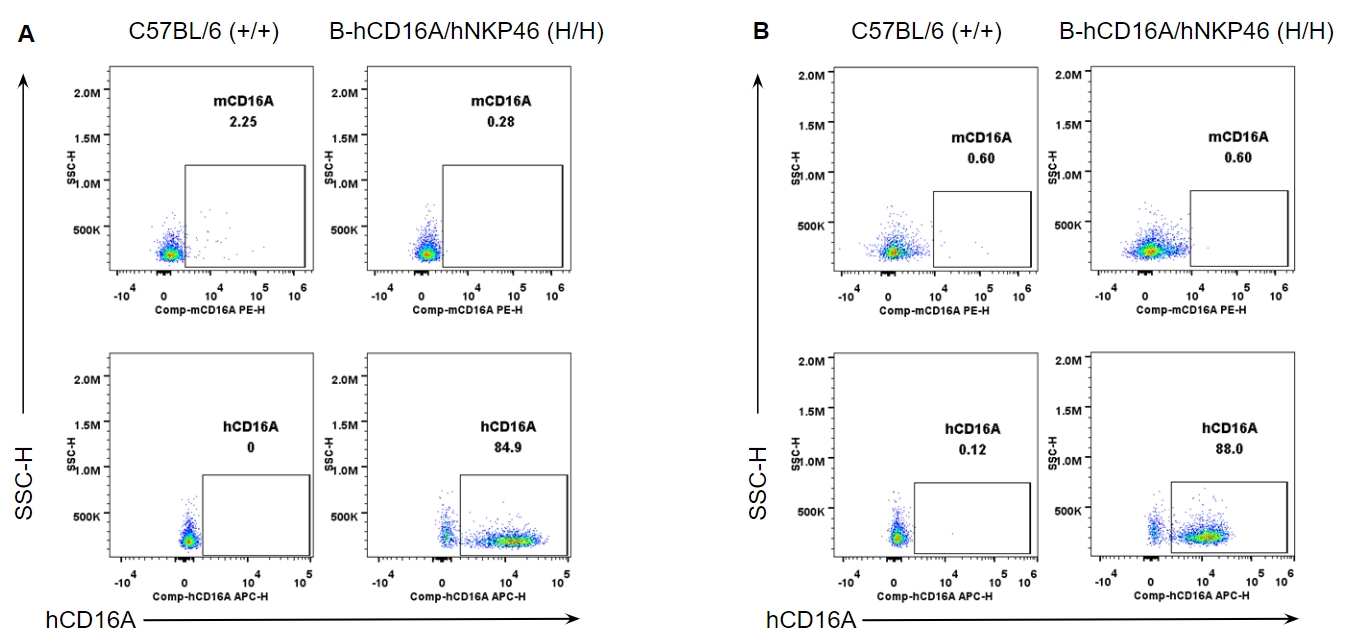
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was not detectable in wild-type mice. Human CD16A was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-macrophages

-
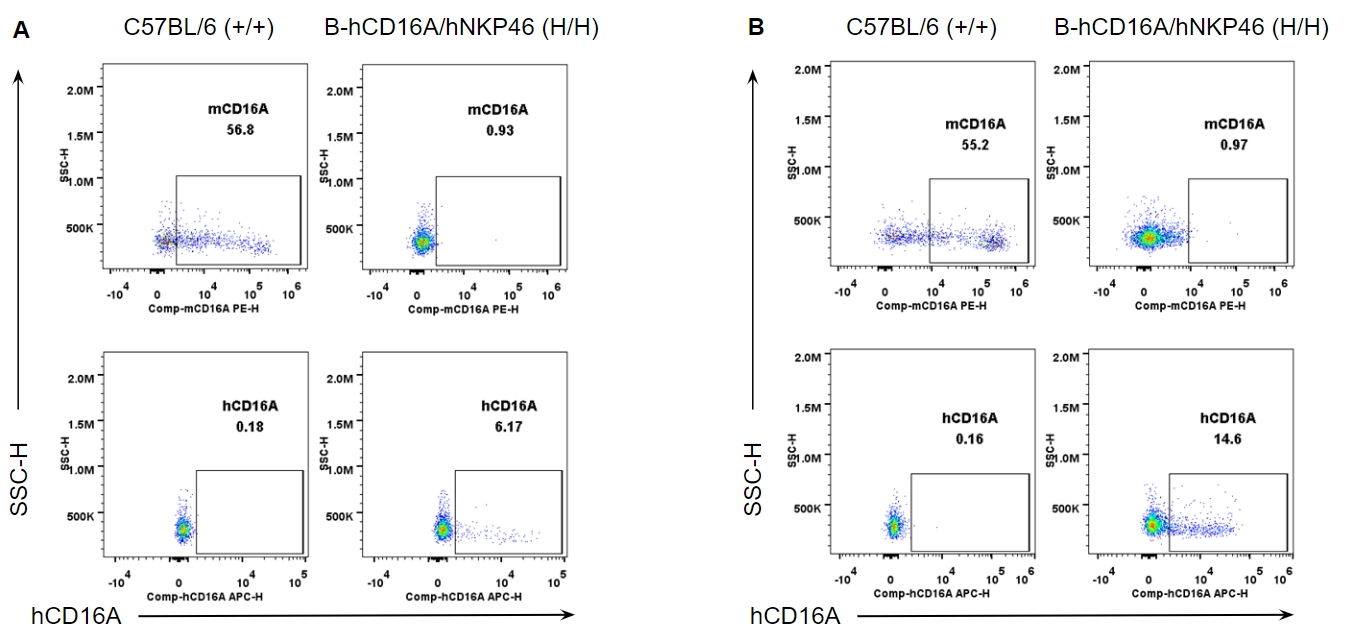
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was detectable in wild-type mice. Human CD16A was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-Peritoneal exudative macrophages(PEMs)

-
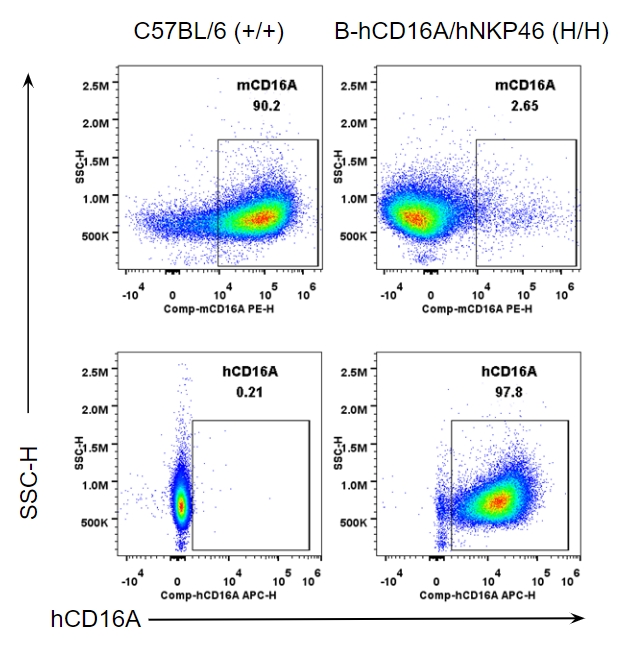
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Peritoneal exudative macrophages(PEMs) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was detectable in wild-type mice. Human CD16A was exclusively detectable in homozygous B-hCD16A/hNKP46 mice.
-
Protein expression analysis of CD16A-granulocytes

-
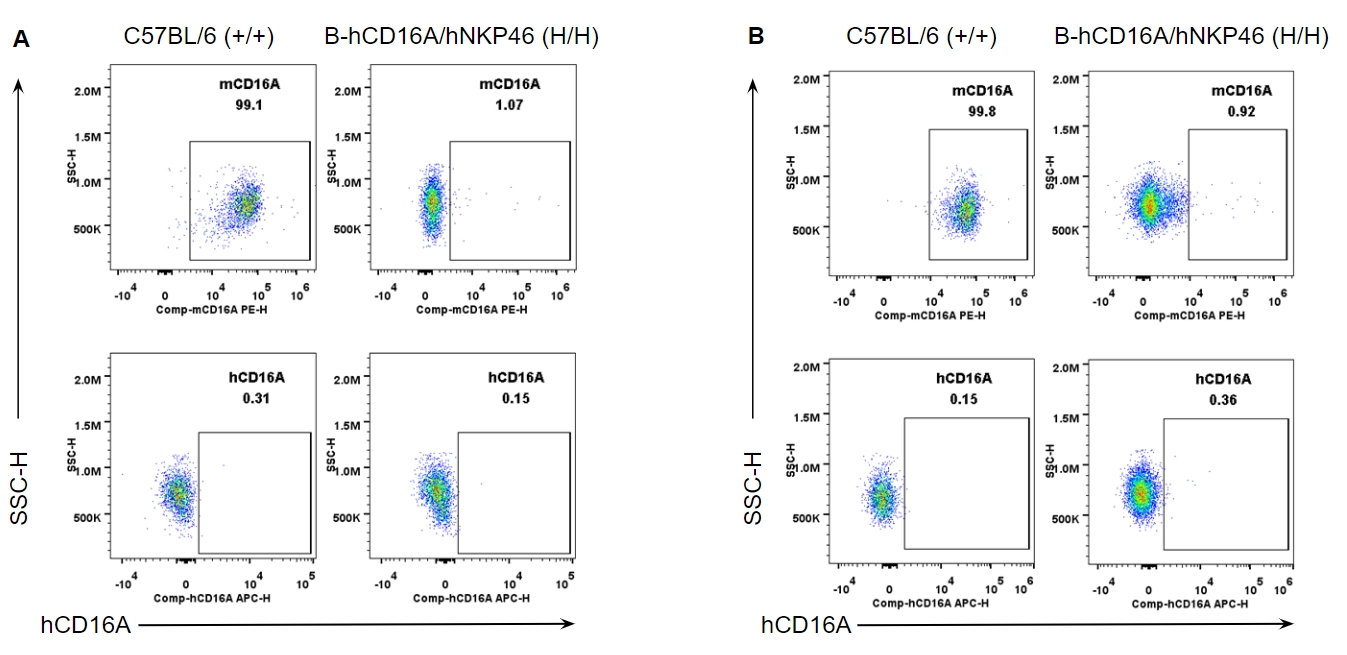
Strain specific CD16A expression analysis in wild-type and B-hCD16A/hNKP46 mice by flow cytometry. Splenocytes (A) and blood cells (B) were collected from wild-type C57BL/6 and homozygous B-hCD16A/hNKP46 (H/H) mice, and analyzed by flow cytometry with species-specific anti-CD16A antibody. Mouse CD16A was detectable in wild-type mice. Human CD16A was not detectable in homozygous B-hCD16A/hNKP46 mice.
-
Analysis of leukocytes cell subpopulation in spleen

-
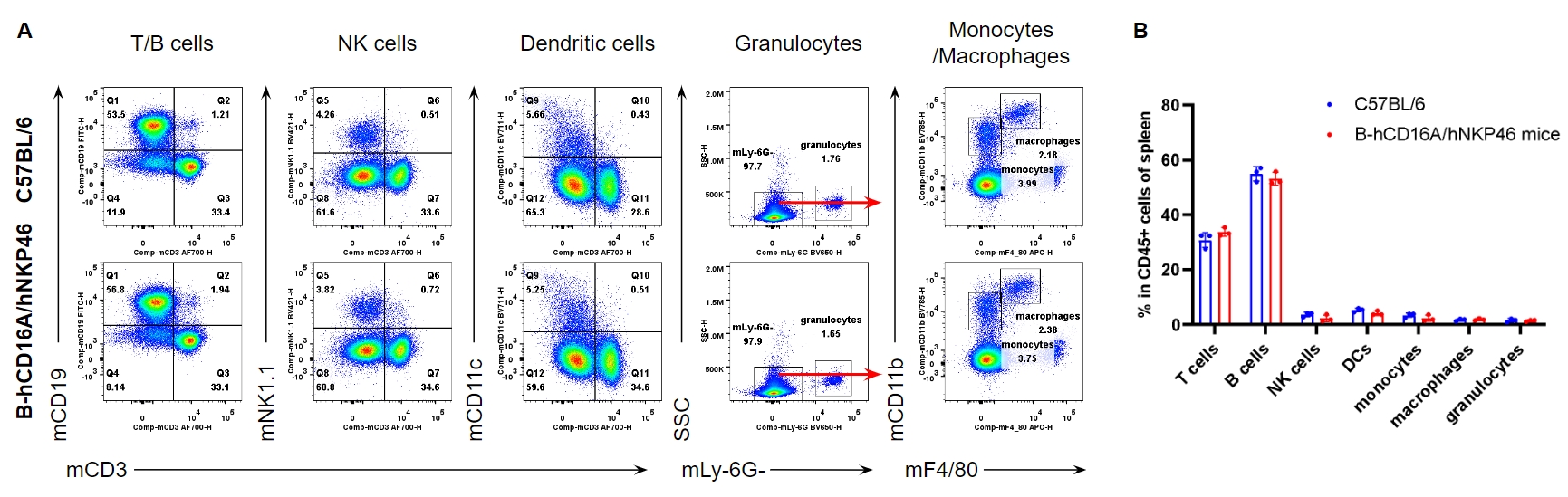
Analysis of spleen leukocyte subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these cell types in spleen. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in spleen

-
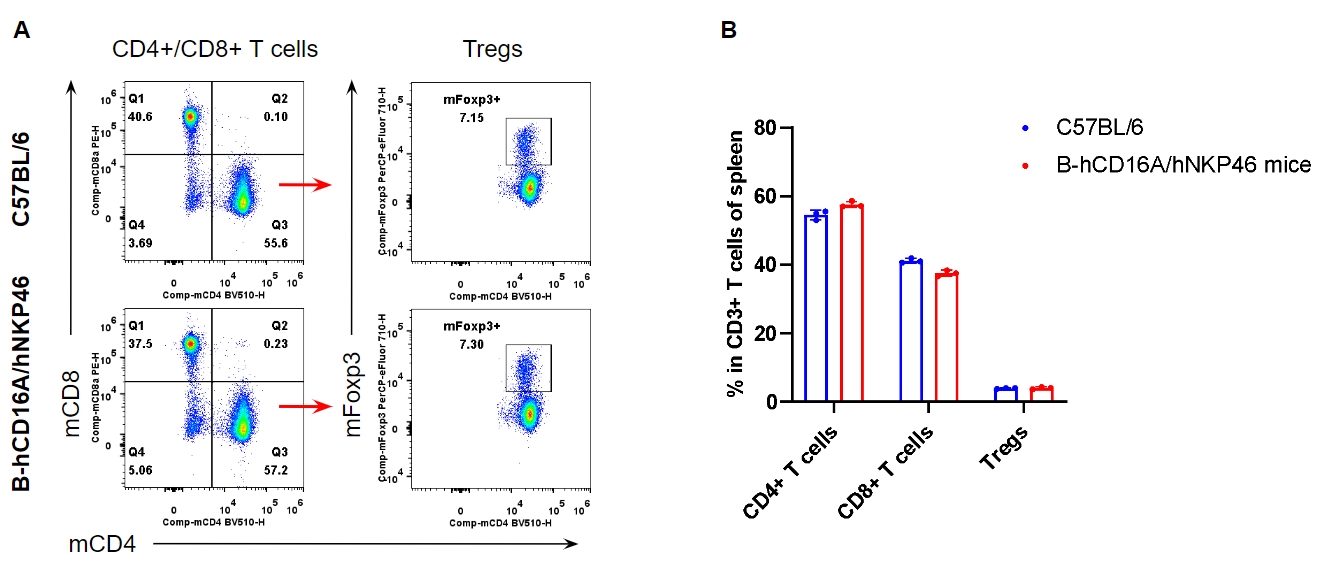
Analysis of spleen T cell subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these T cell subtypes in spleen. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in lymph node

-
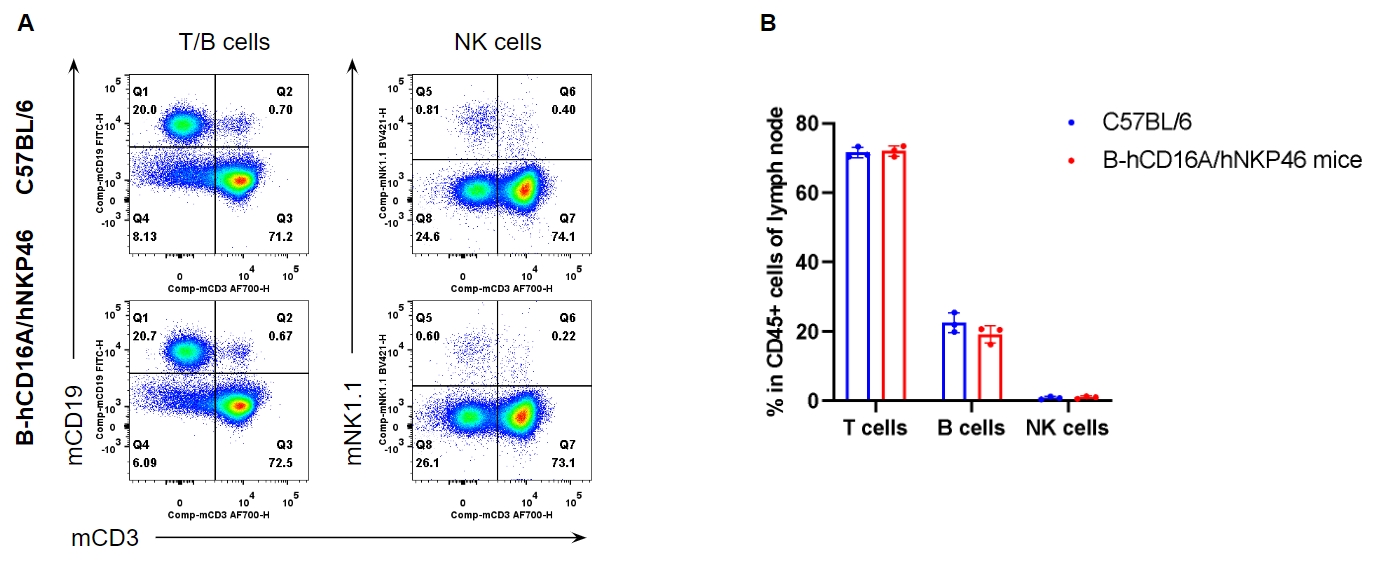
Analysis of lymph node leukocyte subpopulations by FACS. Leukocytes were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the leukocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of T cells, B cells and NK cells in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these cell types in lymph node. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in lymph node

-
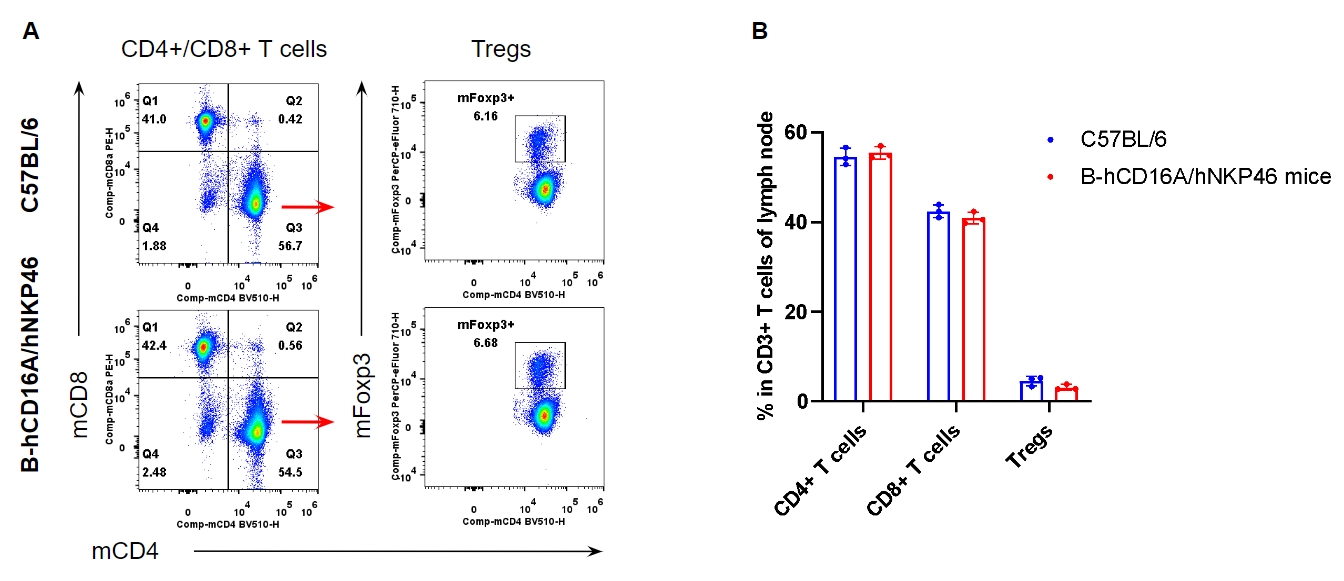
Analysis of lymph node T cell subpopulations by FACS. Leukocytes were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these T cell subtypes in lymph node. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in blood

-
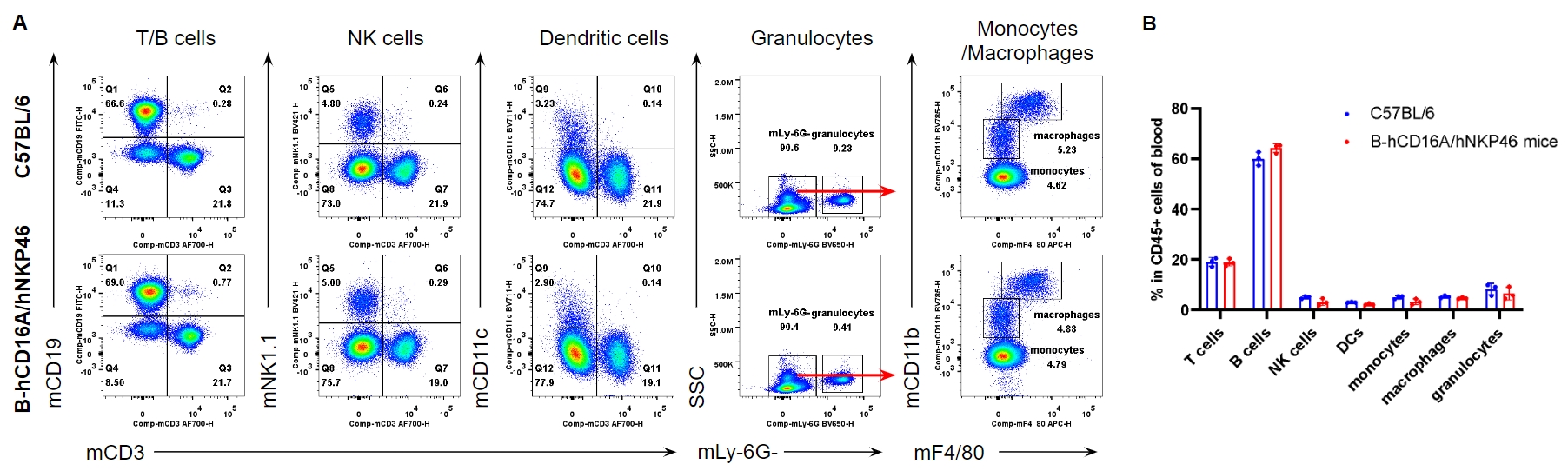
Analysis of blood leukocyte subpopulations by FACS. Blood were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these cell types in blood. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in blood

-
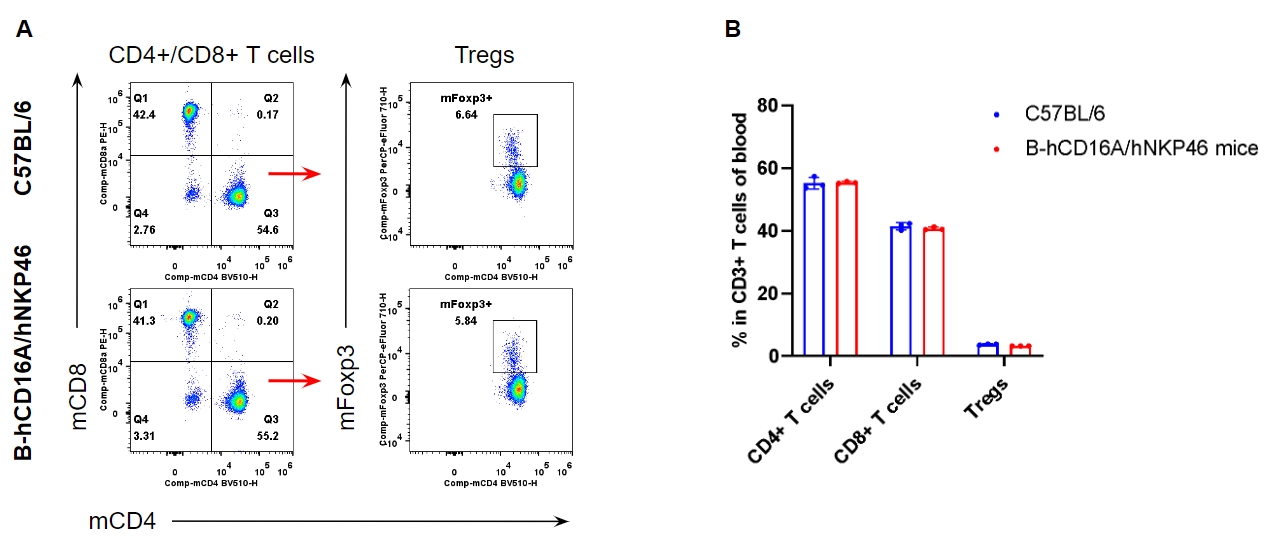
Analysis of blood T cell subpopulations by FACS. Blood were isolated from female C57BL/6 and homozygous B-hCD16A/hNKP46 mice (n=3, 7-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hCD16A/hNKP46 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hCD16A and hNKP46 in place of their mouse counterparts does not change the overall development, differentiation or distribution of these T cell subtypes in blood. Values are expressed as mean ± SEM.
-
Tumor growth curve & Body weight changes

-
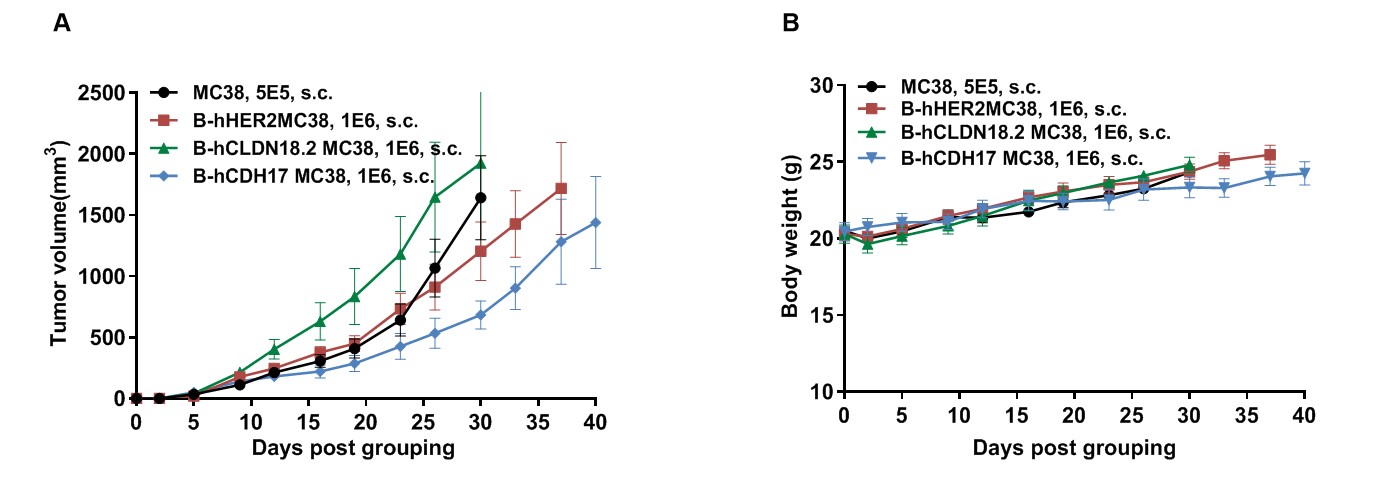
Subcutaneous homograft tumor growth of B-hHER2 MC38 cells, B-hCLDN18.2 MC38 cells, and B-hCDH17 MC38 cells in B-hCD16A/hNKP46 mice. B-hHER2 MC38 cells, B-hCLDN18.2 MC38 cells, and B-hCDH17 MC38 cells (1×106) and wild-type MC38 cells (5×105) were subcutaneously implanted into B-hCD16A/hNKP46 mice (female, 8-week-old, n=6). Tumor volume and body weight were measured twice a week. (A) Average tumor volume ± SEM. (B) Body weight (Mean± SEM). Volume was expressed in mm3 using the formula: V=0.5 X long diameter X short diameter2. As shown in panel A B-hHER2 MC38 cells, B-hCLDN18.2 MC38 cells, and B-hCDH17 MC38 cells were able to establish tumors in B-hCD16A/hNKP46 mice and can be used for efficacy studies.


