Basic Information
Strain Name
C57BL/6-Tcf21tm1(iCre/ERT2)/Bcgen
Stock Number
110141
Common Name
B-Tcf21-iCreERT2 mice
Source/Investigator
Biocytogen Jiangsu Co., Ltd
NCBI Gene ID
Background
C57BL/6
Appearance
Black
Description
This Tcf-iCre/ERT2 model is an efficient tool to study various gene functions when crossed with mice with different loxP site-flanked genes of interest, especially in studies of natural mesenchyme-derived tissue.
-
Details

-
Genotyping PCR Primer
Primer Sequence (5’-3’) Tm (℃) Product Size (bp) Primer-WT-F GACTGCCAGATCCCACCTCAAAC 62 WT:415bp Primer-WT-R GGAGAAGGCCTTGCTCAGCAC 63 Primer-WT-F GACTGCCAGATCCCACCTCAAAC 62 Mut:265bp Primer–Mut-R GCACACAGACAGGAGCATCTTC 59 PCR Reaction System
Reaction Component Volume(µL) Final Concentration ddH2O 14 – 10×Taq buffer 2 1× 10 mM dNTPs 0.5 250 mM each 10 mM Primer-F 0.5 0.25 mM 10 mM Primer-R 0.5 0.25 mM 2.5 U/mL Taq DNA Polymerase Polymerase 0.5 0.0625 U/mL 100-200 ng/mL Template DNA 2 10-20 ng/mL PCR Steps
Step Temp. Time Cycles 1 95℃ 5 min 1 2 95 ℃ 30 s 30 3 61 ℃ 30 s 4 72 ℃ 1 kb / min 5 72 ℃ 10 min 1 6 4℃ hold 1 Determination of Genotypes
Identification of Genotypes According to PCR Results
Primer-WT-F/Primer-WT-R Primer-WT-F /Primer-Mut-R Genotype Mutation Wild Type Mutation N* N Y Mut/Mut N* Y Y Mut/+ N Y N +/+ N*: PCR product was too long to detect Mut/Mut: representing homozygous
N: No expected PCR product detected Mut/+: hemizygous
Y: Expected PCR product detected. +/+: wild type alleleAgarose Gel Electrophoresis
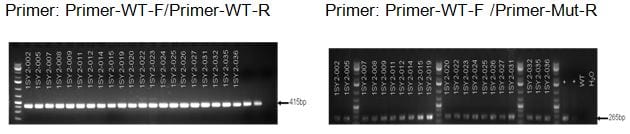
Gene Editing Results
Number Strain Number Genotype 1 1SY2-009 Mut/ + 2 1SY2-011 Mut/ + 3 1SY2-012 Mut/ + 4 1SY2-014 Mut/ + 5 1SY2-015 Mut/ + 6 1SY2-019 Mut/+ 7 1SY2-020 Mut/ + 8 1SY2-022 Mut/ + 9 1SY2-026 Mut/ + 10 1SY2-031 Mut/ + 11 1SY2-032 Mut/ + -
References

-
- Chaushu S; Wilensky A; Gur C; Shapira L; Elboim M; Halftek G; Polak D; Achdout H; Bachrach G; Mandelboim O. 2012. Direct recognition of Fusobacterium nucleatum by the NK cell natural cytotoxicity receptor NKp46 aggravates periodontal disease. PLoS Pathog 8(3):e1002601. [PubMed:22457623]
- Elboim M; Gazit R; Gur C; Ghadially H; Betser-Cohen G; Mandelboim O. 2010. Tumor immunoediting by NKp46. J Immunol 184(10):5637-44. [PubMed:20404273]
- Ghadially H; Horani A; Glasner A; Elboim M; Gazit R; Shoseyov D; Mandelboim O. 2013. NKp46 regulates allergic responses. Eur J Immunol 43(11):3006-16. [PubMed: 23878025]
- Ghadially H; Ohana M; Elboim M; Gazit R; Gur C; Nagler A; Mandelboim O. 2014. NK Cell Receptor NKp46 Regulates Graft-versus-Host Disease. Cell Rep 7(6):1809-14. [PubMed: 24882008]
- Glasner A; Ghadially H; Gur C; Stanietsky N; Tsukerman P; Enk J; Mandelboim O. 2012. Recognition and Prevention of Tumor Metastasis by the NK Receptor NKp46/NCR1. J Immunol 188(6):2509-15. [PubMed: 22308311]
- Gur C; Porgador A; Elboim M; Gazit R; Mizrahi S; Stern-Ginossar N; Achdout H; Ghadially H; Dor Y; Nir T; Doviner V; Hershkovitz O; Mendelson M; Naparstek Y; Mandelboim O. 2010. The activating receptor NKp46 is essential for the development of type 1 diabetes. Nat Immunol 11(2):121-8. [PubMed: 20023661]
- 7. Wang F, Meng M, Mo B, et al. Crosstalks between mTORC1 and mTORC2 variagate cytokine signaling to control NK maturation and effector function[J]. Nature communications, 2018, 9(1): 4874


