Preclinical Models of Asthma
Asthma is a chronic inflammatory disorder of the airway characterized by variable airflow obstruction, airway hyperresponsiveness (AHR) and airway inflammation. The main clinical symptoms of asthma are shortness of breath, wheezing, coughing, and increased mucus secretions upon exposure to allergens. The pathogenesis of asthma is caused by complex interactions between genetic, epigenetic, and environmental factors. Pathologic changes are mediated by several types of airway cells and immune cells, including airway epithelial cells, eosinophils, and T cell subsets. In particular, Th2 cells have been thought to predominate in high eosinophilic asthma, which is characterized by increased levels of IL-4, IL-5, and IL-13.
Biocytogen provides robust and validated asthma models that can be induced by various means. In our OVA model, mice are sensitized by multiple intraperitoneal injections of ovalbumin and then challenged by inhalation of aerosolized ovalbumin. The HDM (house dust mite) and alternaria-induced asthma models involves intranasal challenges spanning a period of 4 weeks. Induction by these means results in increased IgE and eosinophil levels, while histologic staining shows increased airway mucus and inflammatory leukocyte infiltration.
For testing novel therapies targeting anti-human Th2 cytokines implicated in asthma, Biocytogen’s humanized cytokine models can be treated to induce asthma. In particular, we validated the ovalbumin model in B-hTSLP/hTSLPR mice, and the HDM asthma model in B-hIL4/IL4RA mice. In these models, human cytokines and receptors replace their mouse counterparts via genomic knock-in. These mice thus allow direct testing of asthma therapeutics targeting the human IL-4 and/or TSLP pathway in mice.
Watch the webinar: Investigating Inflammatory Disease using Humanized Cytokine Mice
-
Ovalbumin-induced asthma mouse model
-
Establishment of OVA (Ovalbumin)-induced acute asthma model
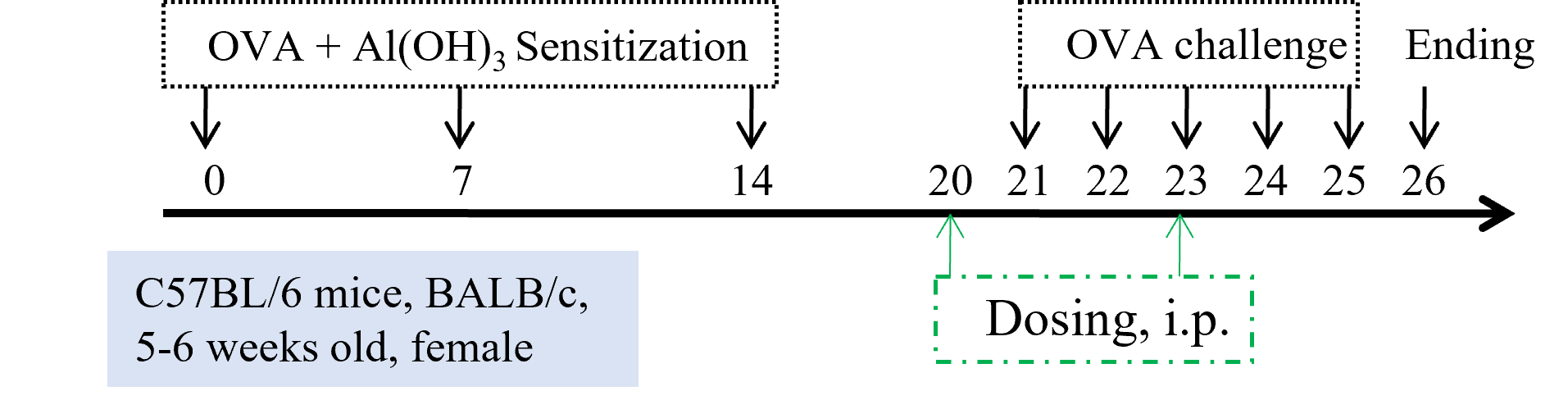
Experimental setup for ovalbumin induction of asthma. After the sensitization period, daily challenge of OVA is provided as indicated by the black arrows. Dosing with test articles can be performed as indicated by the dotted lines.
Immune Cell Infiltration in Bronchoalveolar Lavage Fluid (BALF) and IgE in serum of Asthmatic Mice
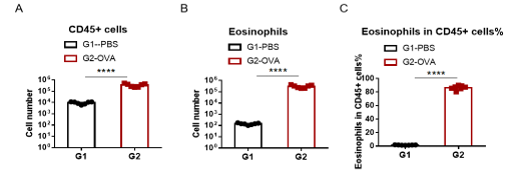
Upon ovalbumin sensitization and challenge, mice showed a considerable increase of infiltration of leukocytes including eosinophils. This represented a clear allergic phenotype at day 26 in BALF when compared with mice not sensitized or challenged.
IgE production in mouse asthma model
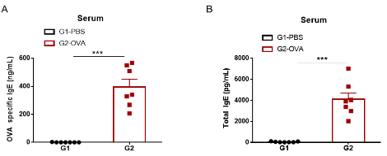
Level of OVA-specific IgE and total IgE antibody were significantly higher in OVA-challenged mice compared with PBS-challenged mice.
H&E staining in mouse asthma model
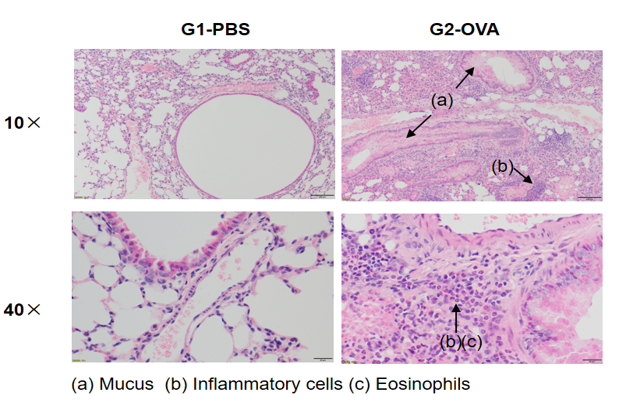
Lung histology of mice of asthmatic mice sensitized and challenged with OVA or PBS. In contrast to the G1 untreated group, the OVA-treated G2 model animals showed asthma-related pathological changes as demonstrated by vascular and peribronchial mixed inflammatory cell infiltration (b) and mucus (a) formation in some bronchi.
Efficacy Validation of Dexamethasone (Dex) in OVA-induced Asthma Mouse Model
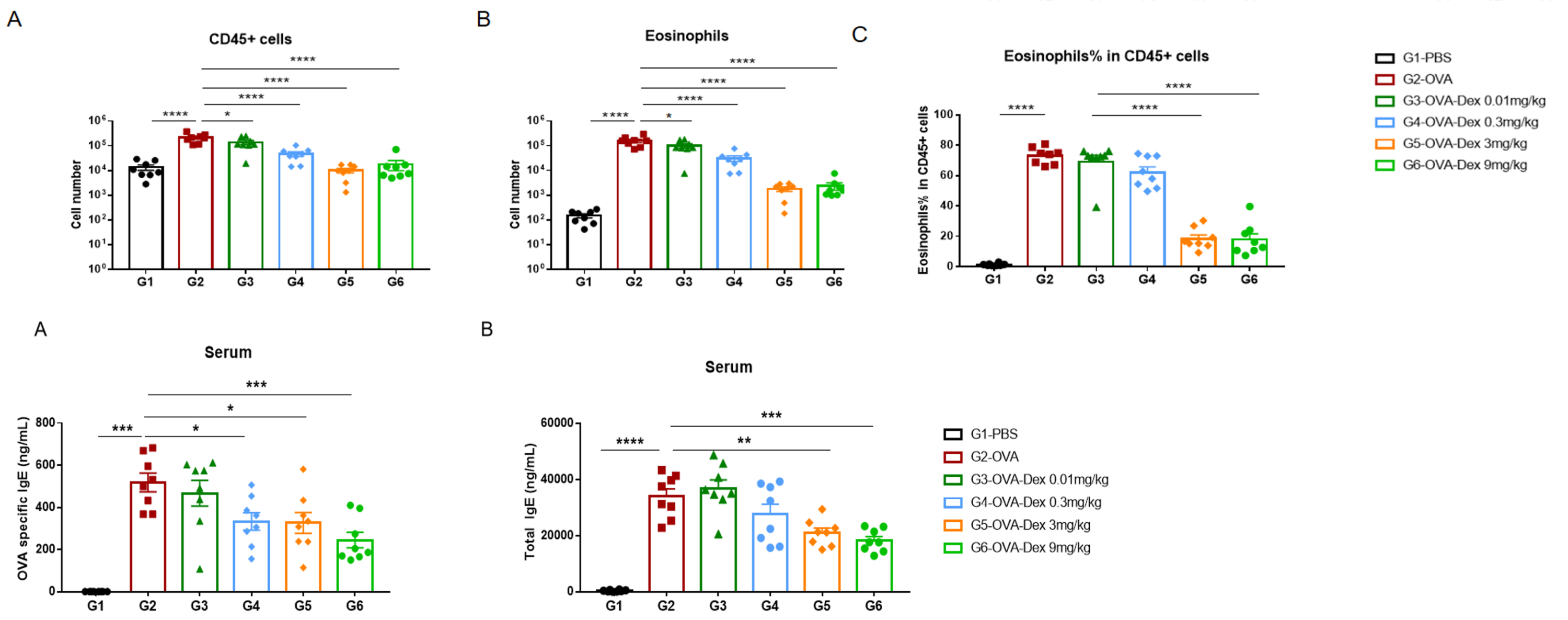
Quantification of immune cells in the BALF (top row) and IgE levels (bottom row) in serum of OVA-induced asthmatic mice. Mice treated with the highest doses of dexamethasone showed a significant reduction in inflammatory infiltration and IgE production.
-
Efficacy of anti-human TSLP in OVA-induced asthma model
-
Establishment of OVA (Ovalbumin)-induced acute asthma model and anti-human TSLP treatment
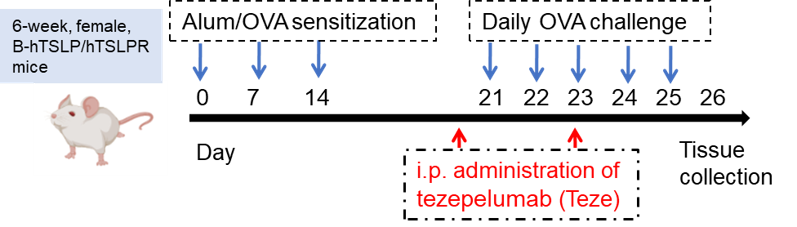
Female B-hTSLP/hTSLPR mice were sensitized and treated with OVA as indicated by the blue arrows. Tezepelumab was administered (i.p.) as indicated by the red arrows.
Immune Cell Infiltration in Bronchoalveolar Lavage Fluid (BALF) and IgE in serum of OVA-induced Asthmatic Mice
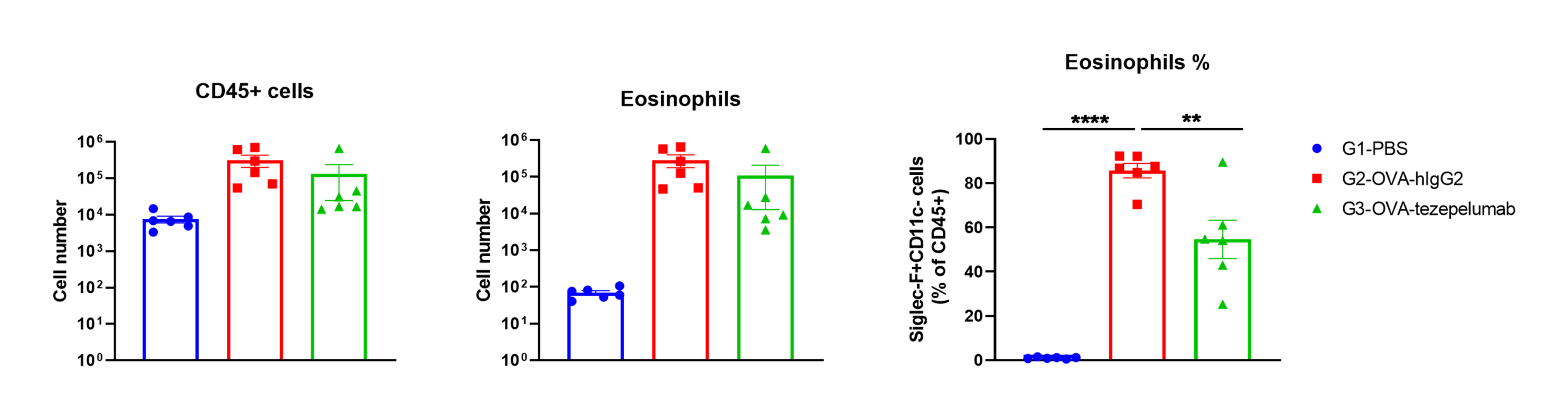
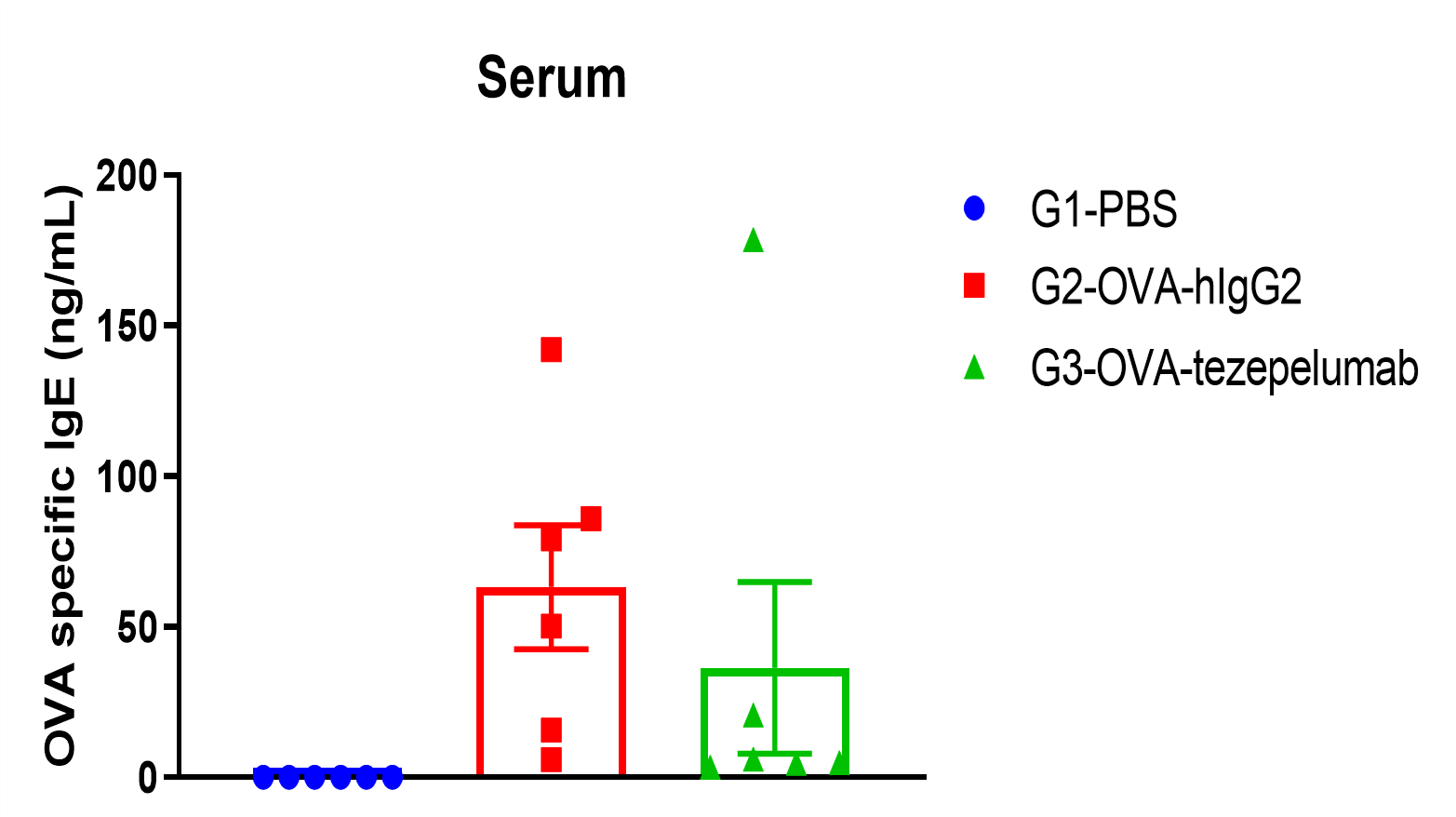
Percentage of eosinophils in mCD45 and total IgE were elevated in serum. The number of CD45+ cells, eosinophils and frequency of eosinophils in the CD45+ cell population after sensitization with OVA. Serum was taken at the experimental endpoint and concentrations of total IgE were measured using ELISA in OVA-induced asthma model in B-hTSLP/hTSLPR mice. Values are expressed as mean ± SEM. *** p<0.001, **p<0.01.
Histologic Assessments in OVA-induced Asthmatic Mice
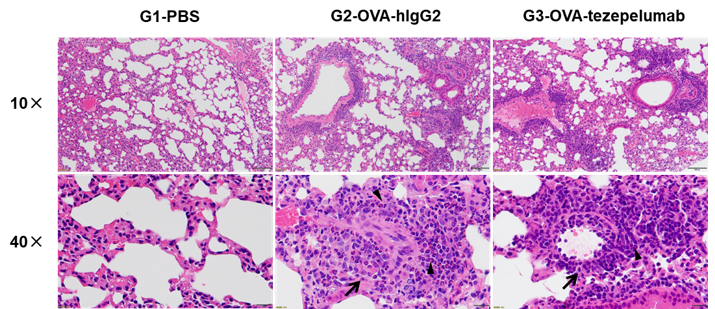
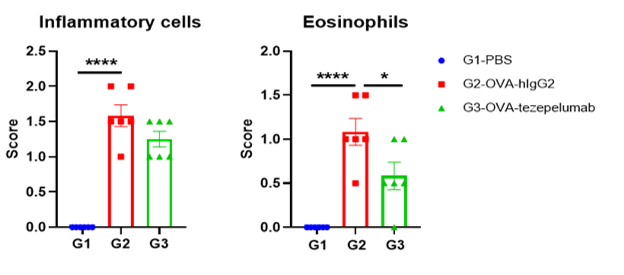
H&E staining of asthma-like model in B-hTSLP/hTSLPR mice plus.
Lung tissues were collected at the study endpoint. H&E staining results revealed that the lung tissues from B-hTSLP/hTSLPR mice plus exposed to PBS aerosols did not show any inflammation. OVA exposure resulted in a significant increase in peribronchial and perivascular inflammation in B-hTSLP/hTSLPR mice plus. A significant reduction in eosinophil infiltration was observed in mice treated with tezepelumab (in house). Black arrow: inflammatory cells; black triangle: eosinophils. -
House dust mite (HDM)-induced asthma model
-
Model Build
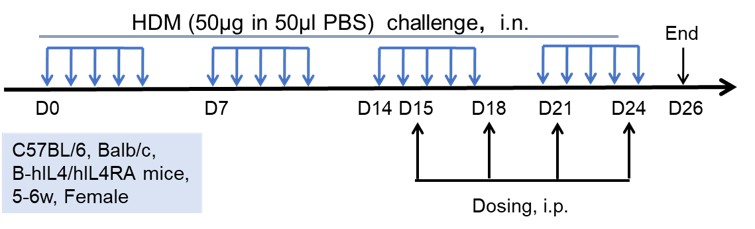
Experimental setup for house dust mite (HDM)-induced asthma. Female mice (C57BL/6, Balb/c, or B-hIL4/IL4RA mice) were subjected to intranasal HDM administration at the timepoints indicated by blue arrows. Dosing with test articles was injected as indicated by the black arrows.
HDM-induced Immune Cell Infiltration in Bronchoalveolar Lavage Fluid (BALF) of Asthmatic Mice
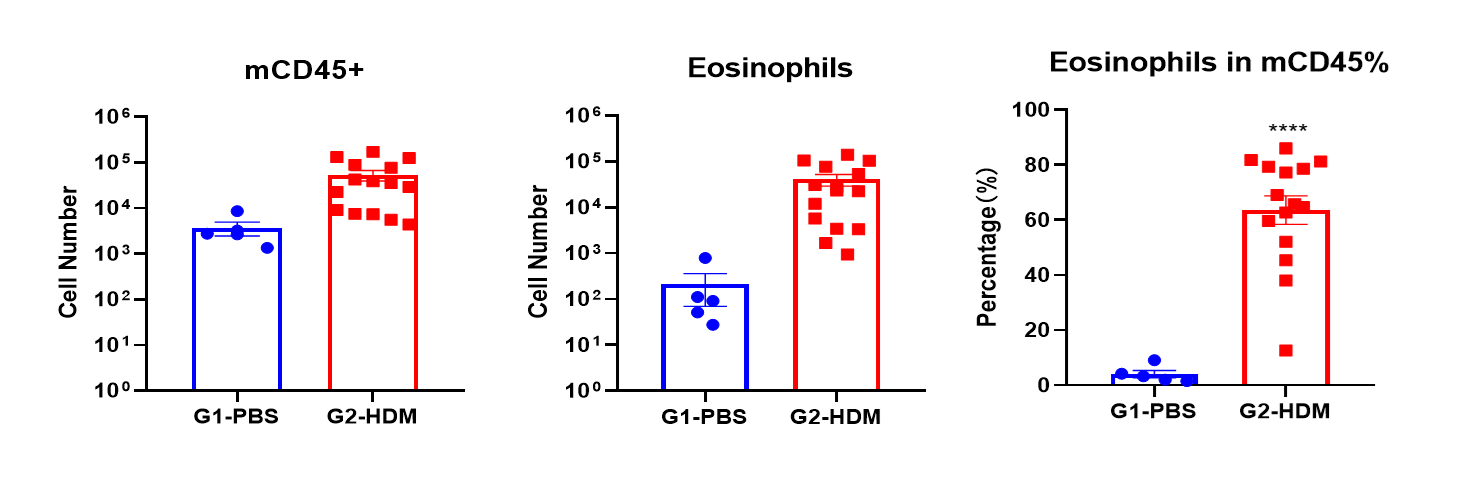
Frequency of CD45+ cells and the number and percentage of eosinophils in the BALF were elevated in mice treated with HDM, as measured using flow cytometry.
HDM-induced Induction of IgE in Serum

Elevated levels of IgE in the serum of mice treated with HDM.
Histologic Assessment of Disease in HDM-induced Asthmatic Mice
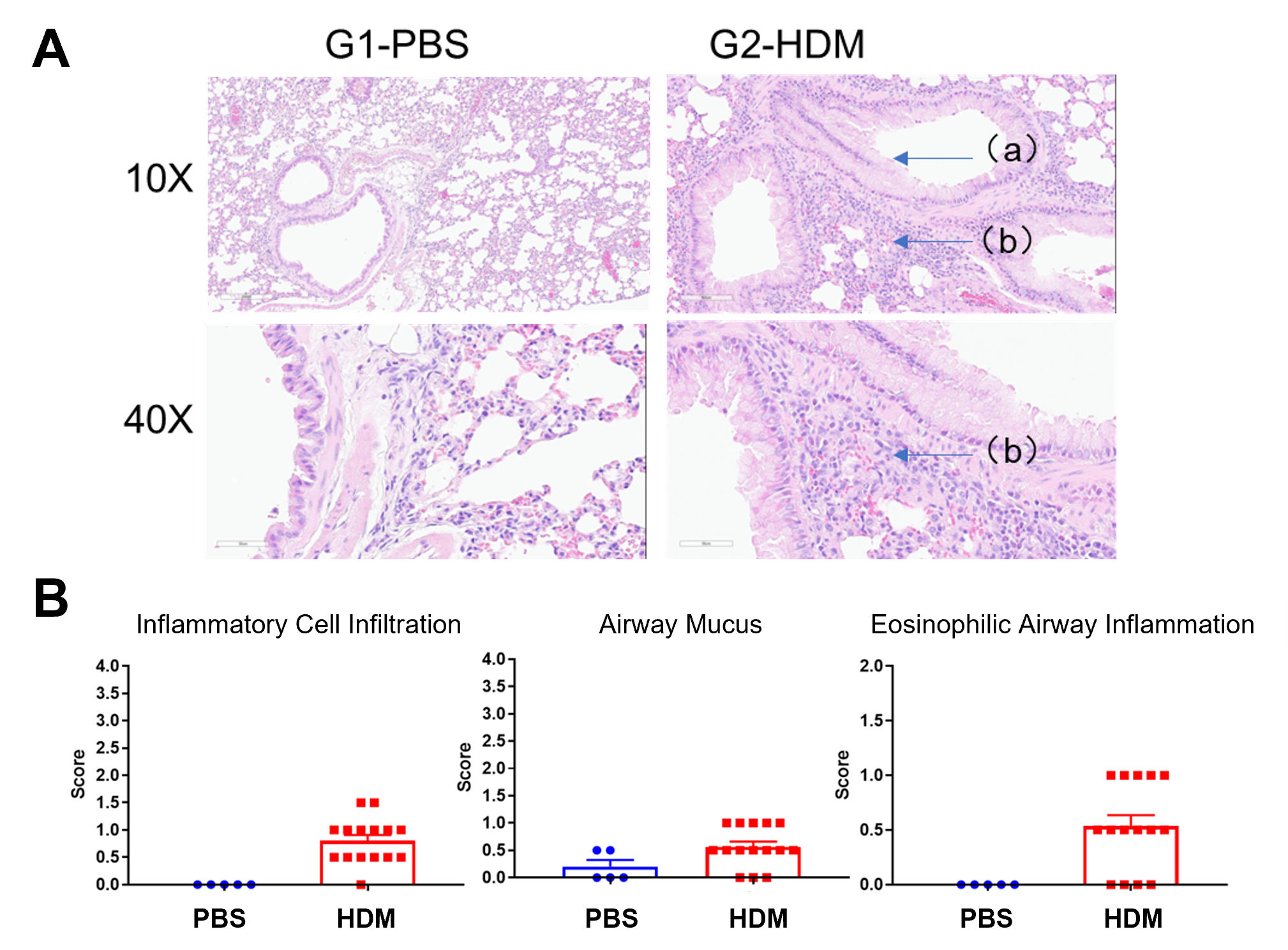
A) H&E sections of the lung in PBS or HDM treated mice. a, mucus; b, inflammatory cell infiltration. B) Histologic scoring of the levels of inflammatory cell infiltration, airway mucus and eosinophilic inflammation in the airway.
Dupilumab Efficacy in B-hIL4/hIL4RA Mice with HDM-induced Asthma
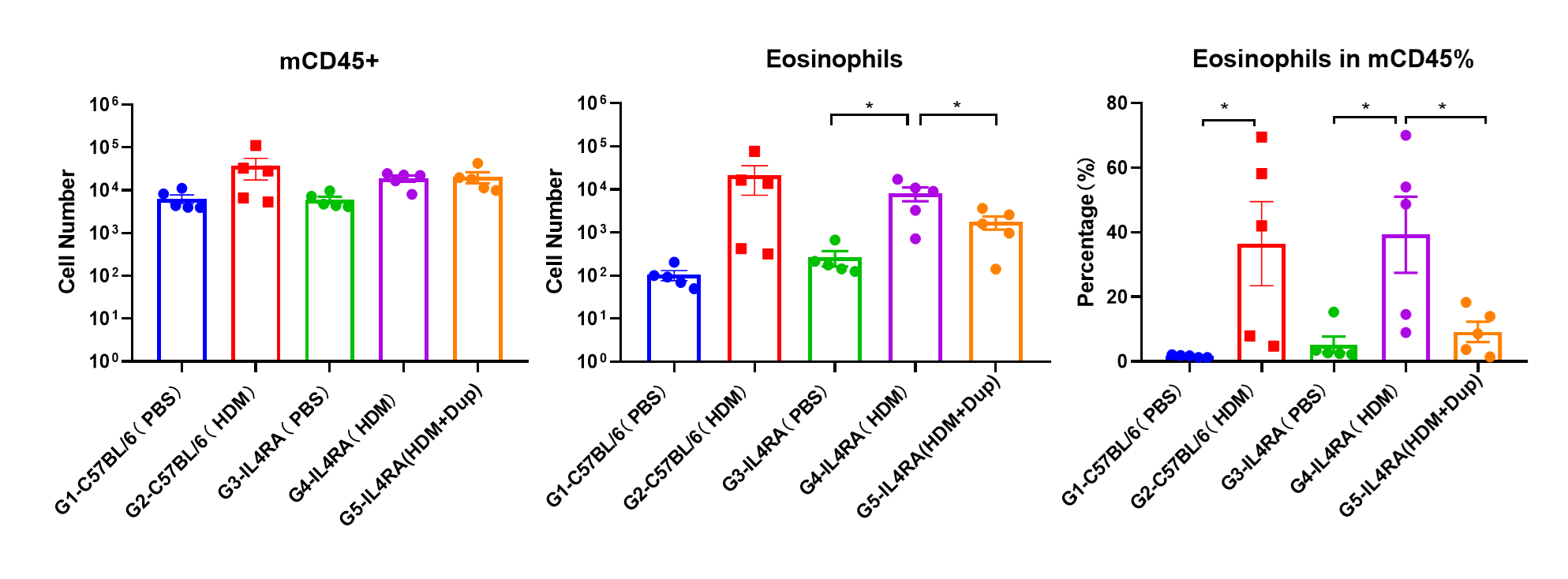
Immune Cell Infiltration in Bronchoalveolar Lavage Fluid (BALF) of Asthmatic Mice. Flow cytometry assessments indicate a significant infiltration of eosinophils in the BALF of HDM-treated mice compared with PBS controls. Dupilumab treatment of HDM-induced mice reduced the number of eosinophils.
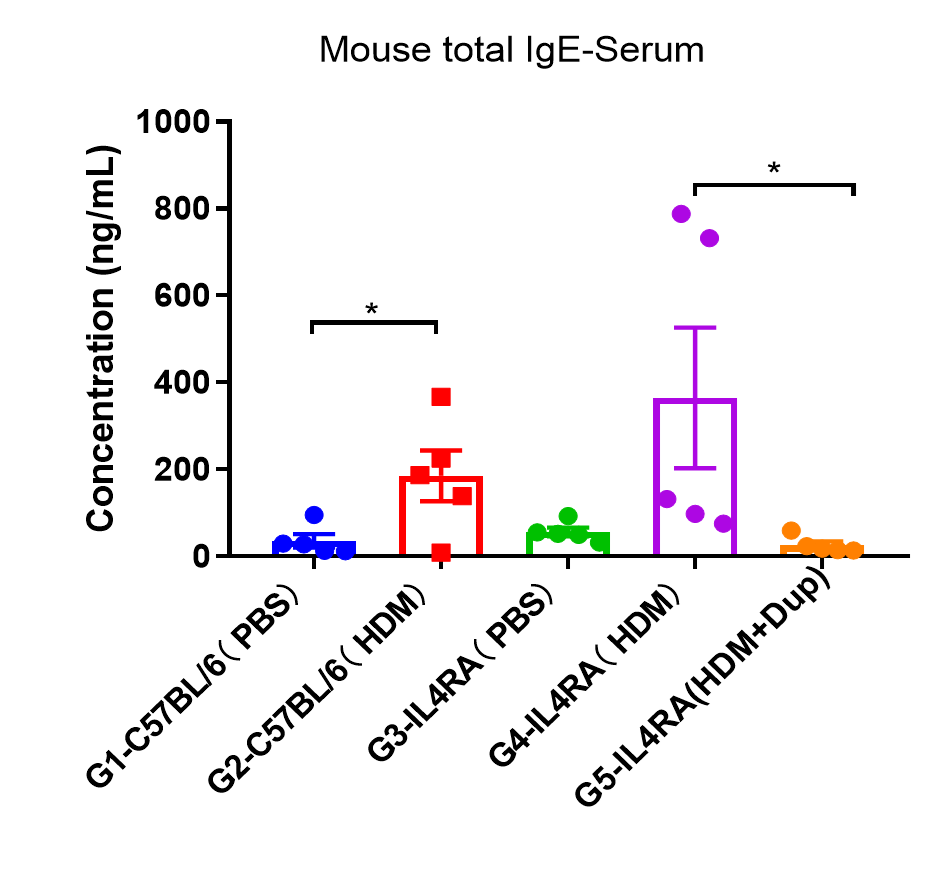
The levels of total IgE in the HDM model group was significantly increased compared with PBS- control group, suggesting successful modeling. Total IgE levels were significantly lower in B-hIL4/hIL4RA mice after administration of dupilumab (in house) (G5) compared with the HDM-only B-hIL4/hIL4RA group (G4). Values are expressed as mean ± SEM. * p<0.05.
-
Alternaria (ALT)-induced asthma mouse model
-
Establishment of Asthma Mouse Model
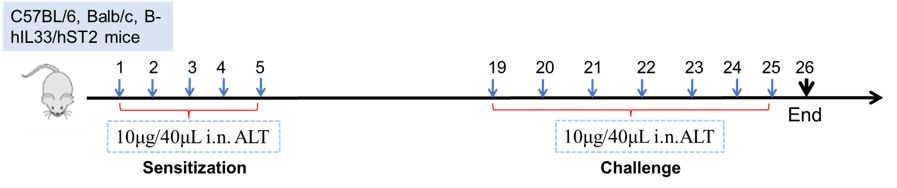
Experimental schematic for alternaria (ALT)-mediated induction of asthma in mice. Mice (C57BL/6, Balb/c, or other humanized strains, such as B-hIL33/hST2 mice), are first sensitized with daily administration of ALT for 5 consecutive days, followed by a challenge from days 19-25 as shown. Bronchioalveolar lavage fluid (BALF), serum, and lungs are assessed for metrics of the disease at the endpoint on day 26.
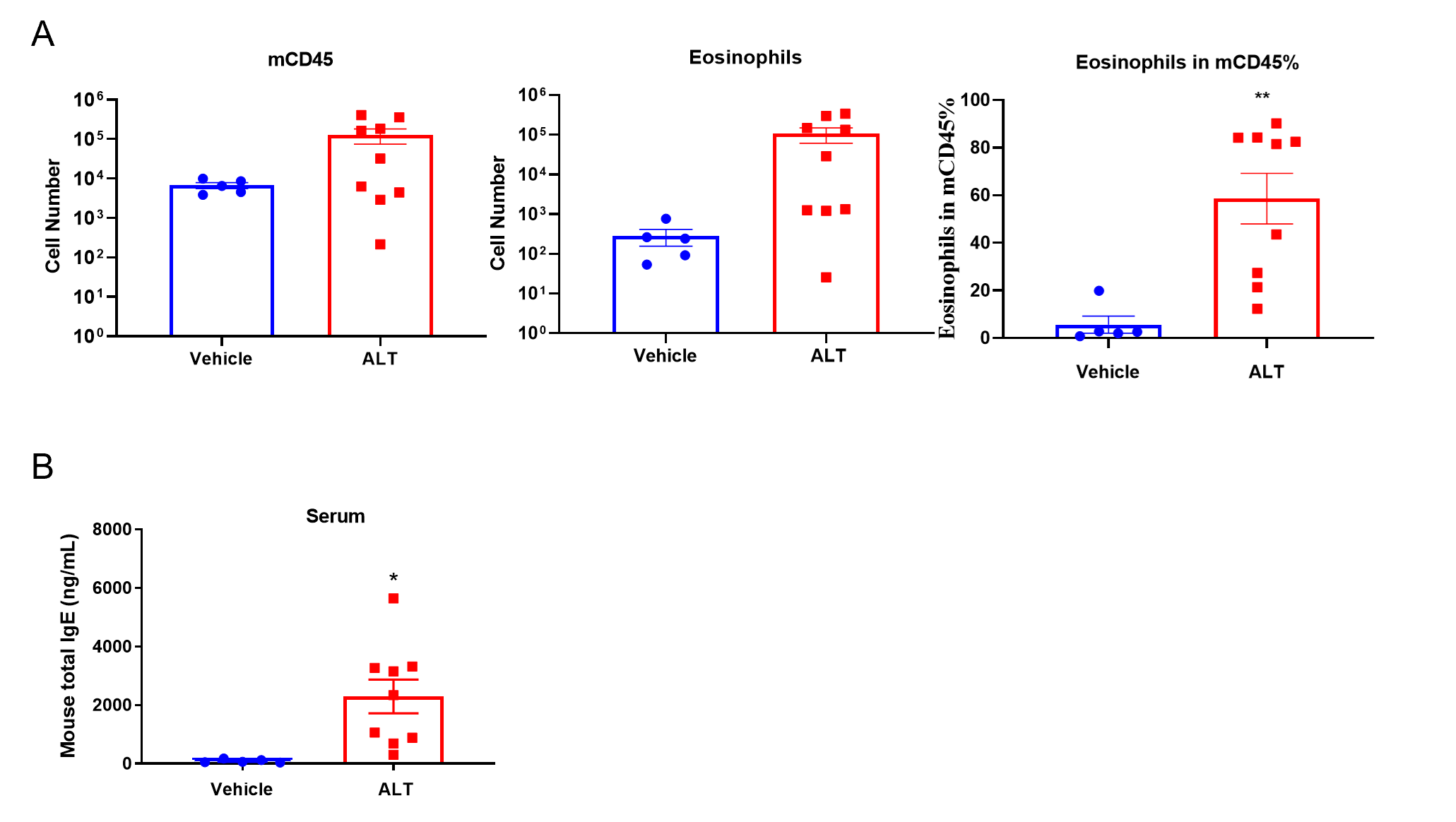
Quantification of Immune Cells in BALF and total IgE levels in serum of ALT-induced Asthmatic mouse model based on C57BL/6 mice. (A) number of CD45+ cells, eosinophils and frequency of eosinophils in the CD45+ cell population after sensitization and challenge with ALT. (B) Serum was taken at the experimental endpoint and concentrations of total IgE were measured using ELISA. Values are expressed as mean ± SEM. *p<0.05, **p<0.01.
Histologic assessment of ALT-treated asthmatic mice
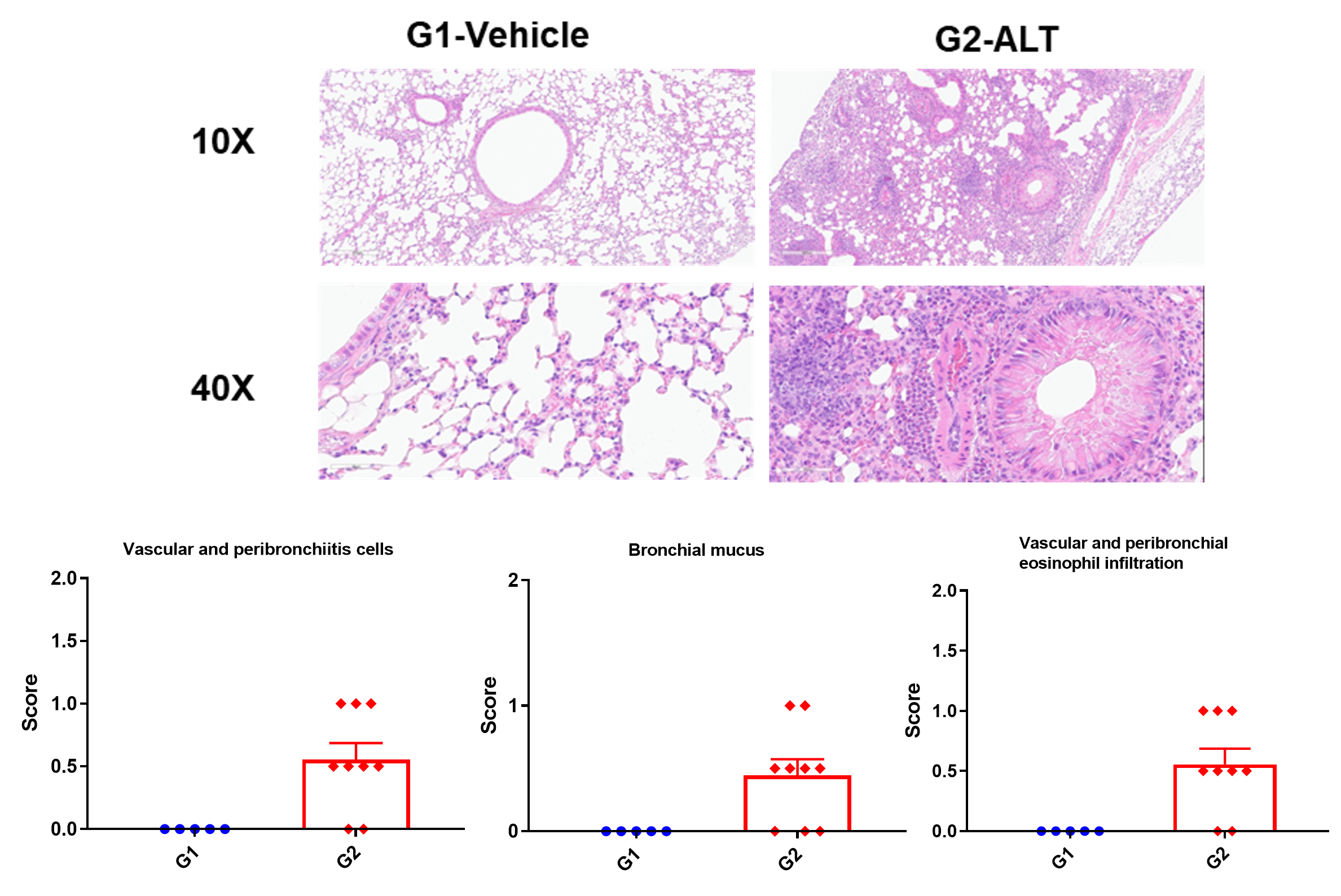
H&E staining in the lungs of asthmatic mice. ALT treatment showed asthma-related pathological changes as demonstrated by vascular and peribronchial mixed inflammatory cell infiltration and mucus formation in some bronchi. The histology scores of inflammatory cells and infiltration of vascular and peribronchial eosinophils were increased after ALT treatment.



