Basic Information
-
Gene targeting strategy

-
Gene targeting strategy for B-hTGFB1 mice. The human TGFB1 gene (encoding the full-length protein) was inserted following part of exon 2 of the mouse Tgfb1 gene to generate B-hTGFB1 mice.
-
mRNA expression analysis

-
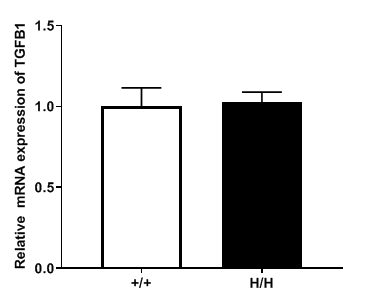
TNFR2 gene expression analysis in wild-type and B-hTGFB1 mice by RT-qPCR. TGFB1 mRNA expression levels in homozygous B-hTGFB1 mice (H/H) was similar to those in wild-type C57BL/6 mice (+/+), demonstrating that introduction of hTGFB1 in place of its mouse counterpart does not change the expression level of TGFB1 mRNA.
-
Protein expression analysis

-
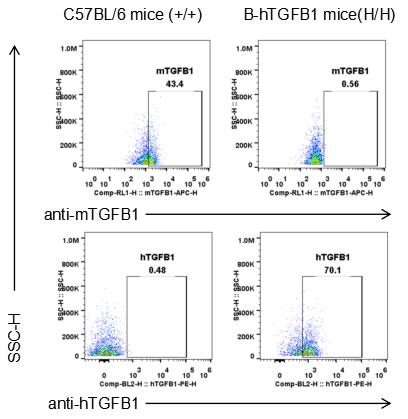
Strain specific TGFB1 expression analysis in wild-type and B-hTGFB1 mice plus by flow cytometry. Splenocytes was collected from wild-type C57BL/6 and homozygous B-hTGFB1 mice, and analyzed by flow cytometry with species-specific TGFB1 antibody. Mouse TGFB1 was detectable in wild-type mice. Human TGFB1 was exclusively detectable in homozygous B-hTGFB1 mice.
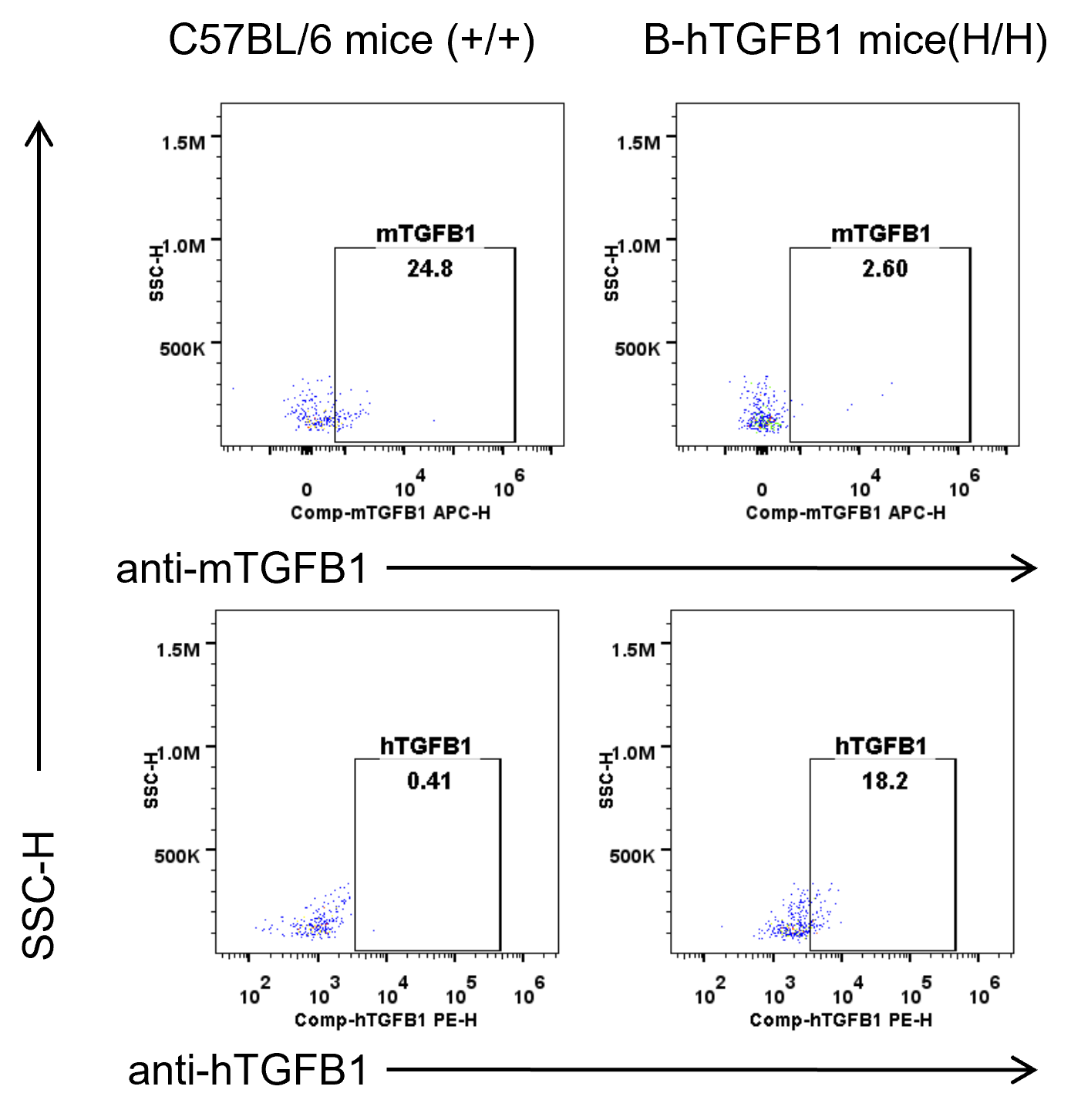
Strain specific TGFB1 expression analysis in wild-type and B-hTGFB1 mice plus by flow cytometry. Splenocytes was collected from wild-type C57BL/6 and homozygous B-hTGFB1 mice, and analyzed by flow cytometry with species-specific TGFB1 antibody. Mouse TGFB1 was detectable in wild-type mice. Human TGFB1 was exclusively detectable in homozygous B-hTGFB1 mice.
-
Analysis of leukocytes in spleen

-
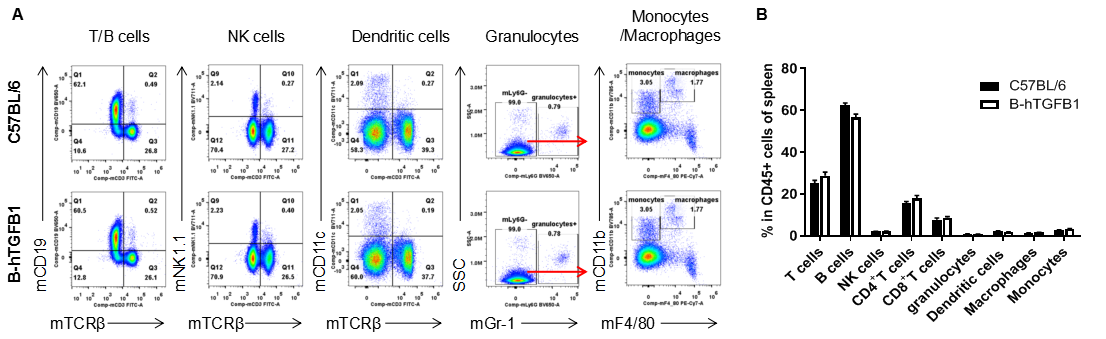
Analysis of spleen leukocyte subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and homozygous humanized B-hTGFB1 mice(n=3, 6-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in homozygous B-hTGFB1 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hTGFB1 in place of its mouse counterpart does not change the overall development, differentiation or distribution of these cell types in spleen. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulations in spleen

-
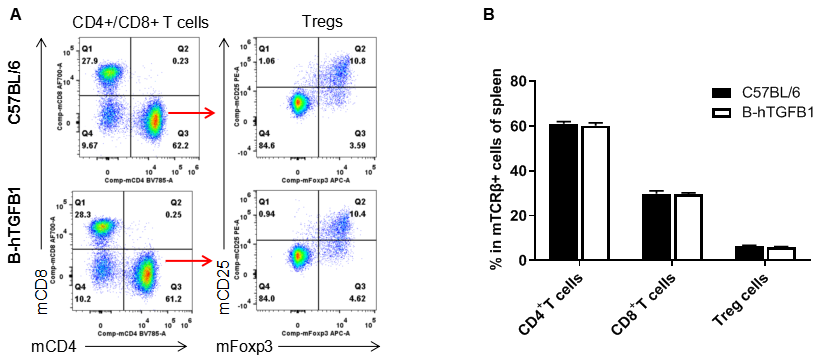
Analysis of spleen T cell subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and homozygous humanized B-hTGFB1 mice (n=3, 6-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for CD3+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD8+ T cells, CD4+ T cells, and Tregs in homozygous B-hTGFB1 mice were similar to those in the C57BL/6 mice, demonstrating that introduction of hTGFB1 in place of its mouse counterpart does not change the overall development, differentiation or distribution of these T cell subtypes in spleen. Values are expressed as mean ± SEM.
-
Platelet Expression

-
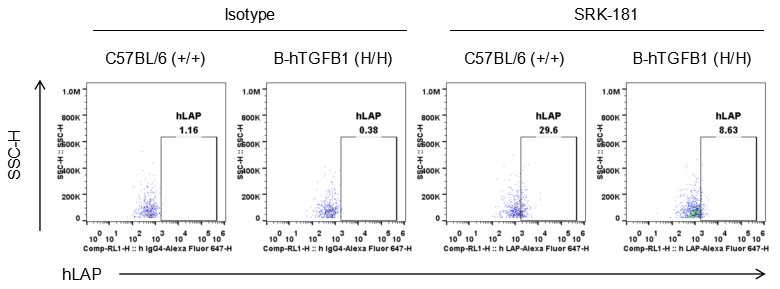
Analysis of platelets in B-hTGFB1 mice by FACS. Blood was isolated from female humanized B-hTGFB1 mice. Flow cytometry analysis of the platelets was performed to assess human LAP expression. LAP was detectable on wild-type C57BL/6 mice and homozygous B-hTGFB1 mice.
-
Combination therapy of anti-mouse PD-1 antibody and anti-human TGFβ1 antibody

-
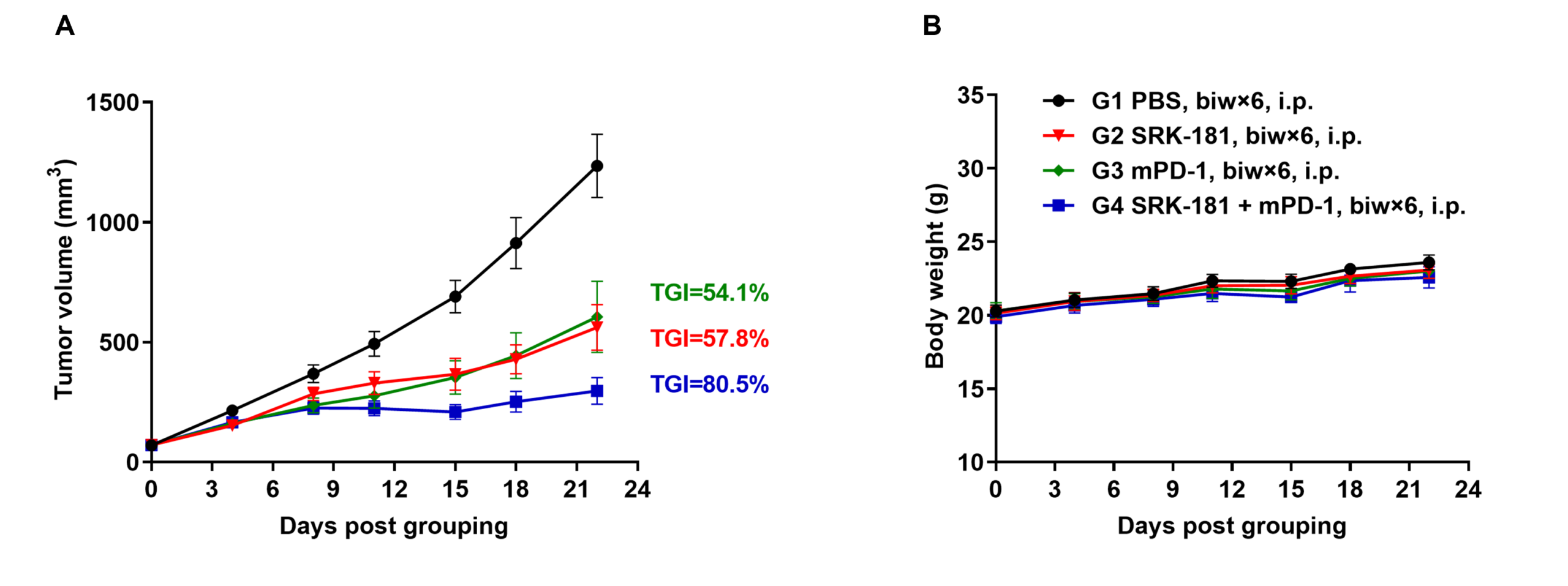
Antitumor activity of anti-mouse PD-1 antibody combined with anti-human TGFβ1 antibody in B-hTGFB1 mice. (A) Anti-mouse PD-1 antibody combined with anti-human TGFβ1 antibody (in house) inhibited MC38 tumor growth in B-hTGFB1 mice. Murine colon cancer MC38 cells (5E5) were subcutaneously implanted into homozygous B-hTGFB1 mice (female, 7-week-old, n=6). Mice were grouped when tumor volume reached approximately 50~80 mm3, at which time they were treated with anti-mouse PD-1 antibody and anti-human TGFβ1 antibody with doses and schedules indicated in panel A. (B) Body weight changes during treatment. As shown in panel A, combination of anti-mPD-1 antibody and anti-human TGFβ1 antibody were efficacious in controlling tumor growth in B-hTGFB1, demonstrating that the B-hTGFB1 mice provide a powerful preclinical model for in vivo evaluation of anti-human TGFB1 antibodies. Values are expressed as mean ± SEM.


