Basic Information
-
Targeting strategy

-
Gene targeting strategy for B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice.
Bacterial artificial chromosome (BAC) clones for the human LILRB1, LILRB2, LILRB3 and LILRB4 genes were integrated into themouse genome randomly.
-
LILRB1 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (non-tumor bearing)

-
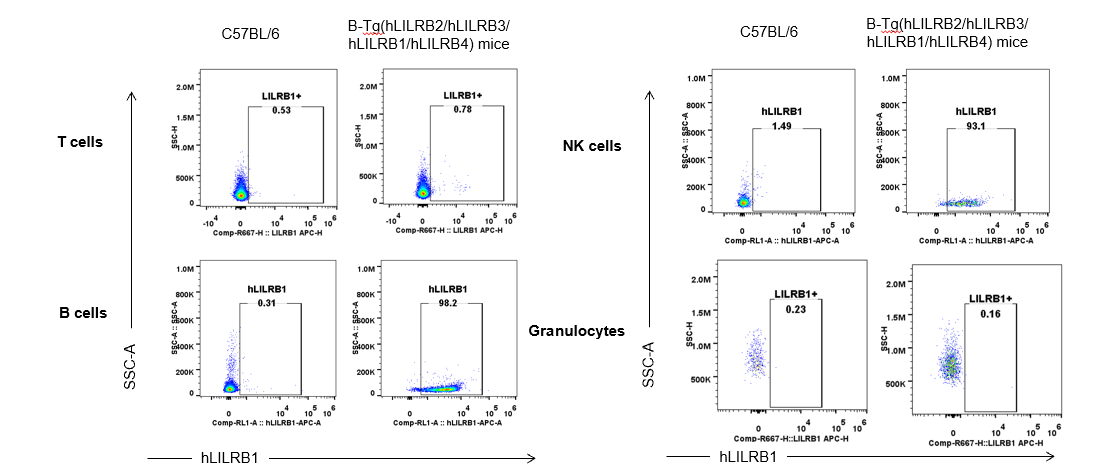
Strain specific hLILRB1 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was both detectable in B cells and NK cells from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
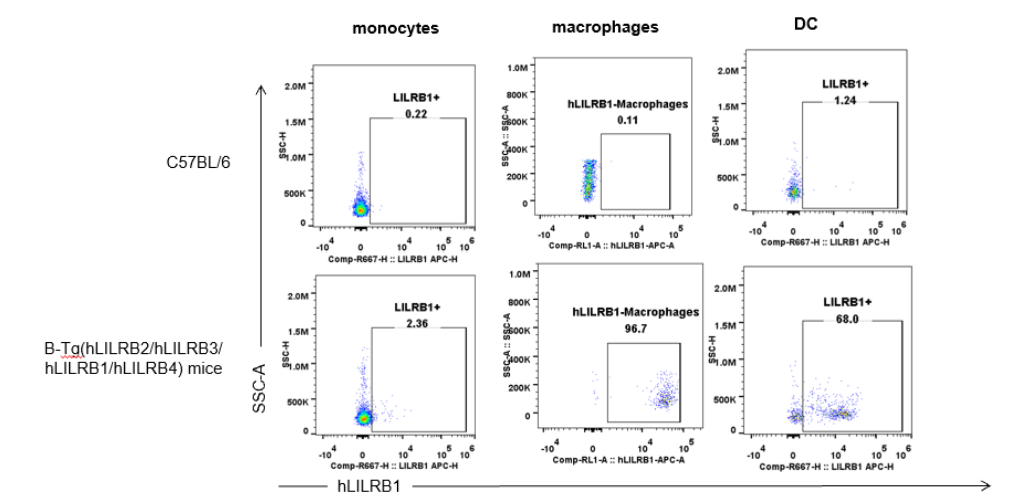
Strain specific hLILRB1 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB1 antibody. Human LILRB1 was both detectable in DCs and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
-
LILRB2 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (non-tumor bearing)

-
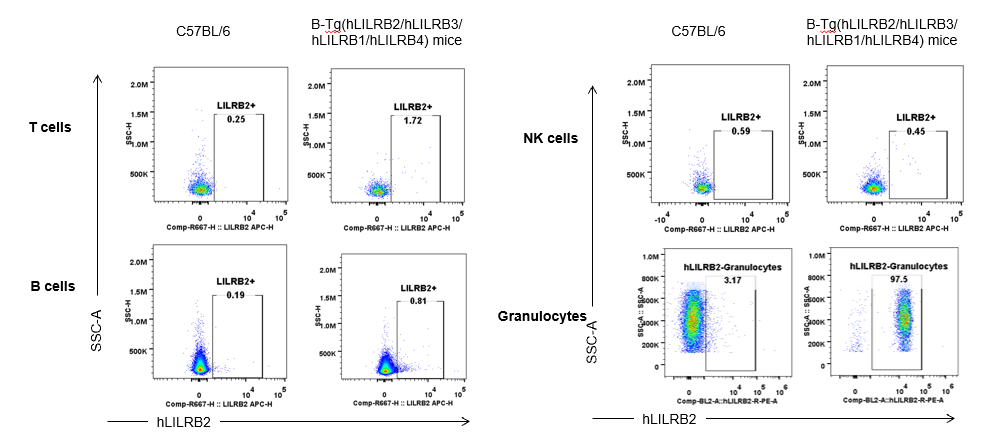
Strain specific hLILRB2 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB2 antibody. Human LILRB2 was detectable in granulocytes from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
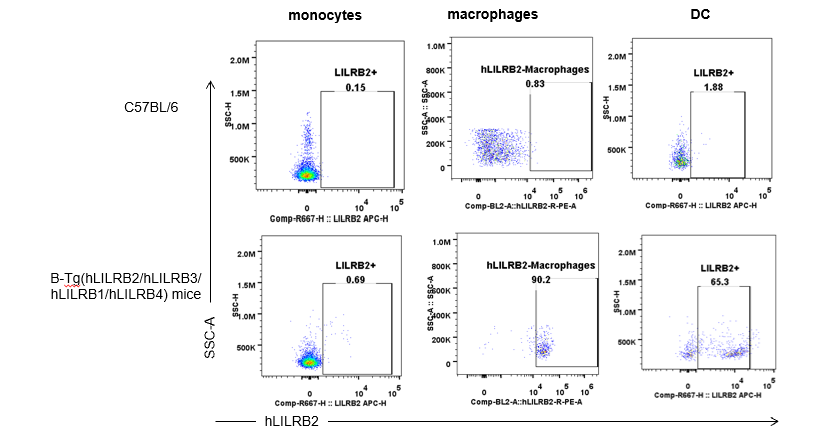
Strain specific hLILRB2 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB2 antibody. Human LILRB2 was both detectable in DCs and macrophages from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
-
LILRB3 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (non-tumor bearing)

-
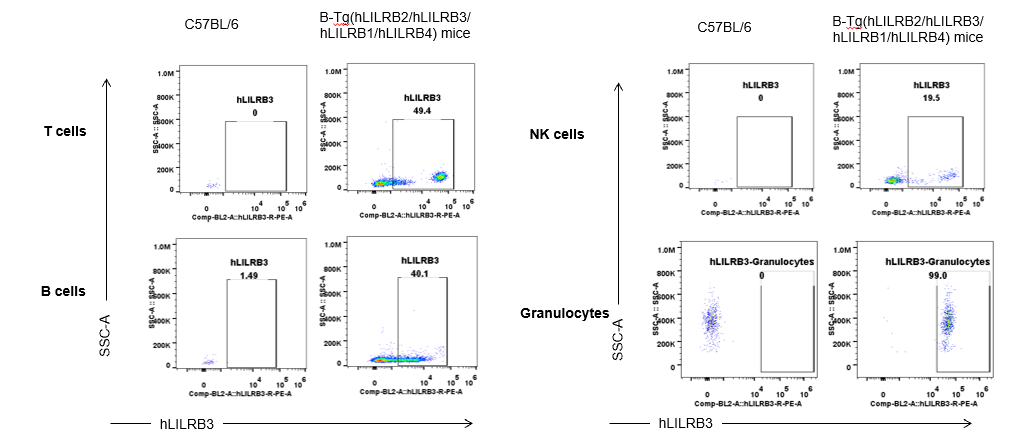
Strain specific hLILRB3 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB3 antibody. Human LILRB3 was both detectable in T cells, B cells, NK cells and granulocytes from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
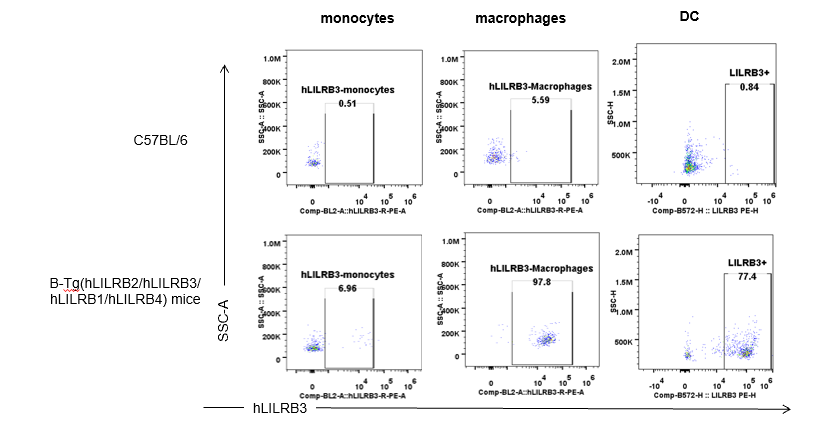
Strain specific hLILRB3 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB3 antibody. Human LILRB3 was both detectable in monocytes, macrophages and DCs from B-Tg(hLILRB1/hLILRB4) mice but not wild-type mice.
-
LILRB4 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (non-tumor bearing)

-
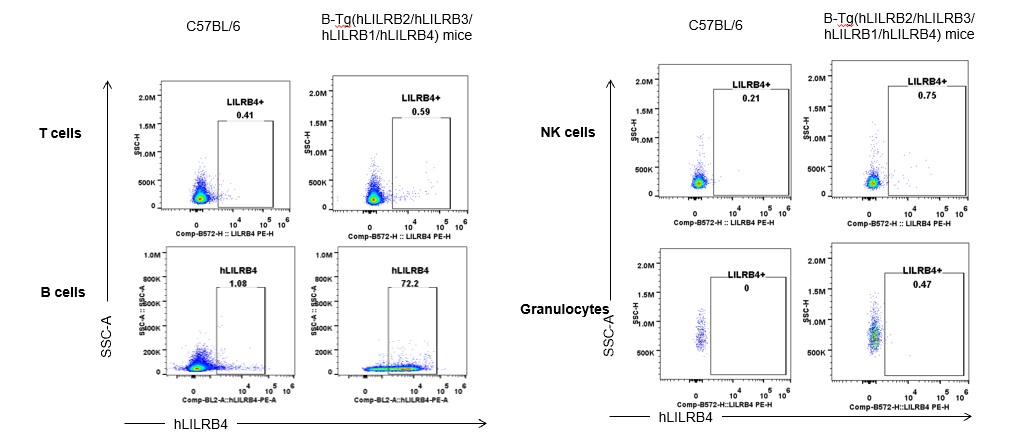
Strain specific hLILRB4 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was detectable in B cells from B-Tg(hLILRB1/hLILRB4) mice.
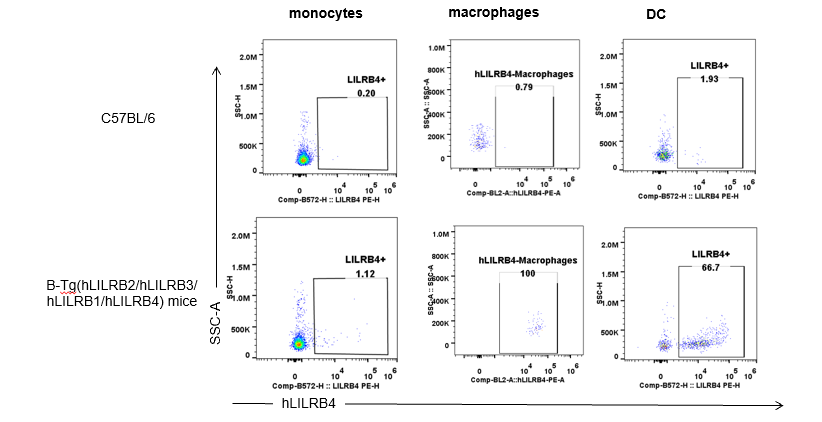
Strain specific hLILRB4 expression analysis in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice by flow cytometry. Blood were collected from wild-type (+/+) and transgenic B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4) mice (Tg), and analyzed by flow cytometry with species-specific anti-human LILRB4 antibody. Human LILRB4 was both detectable in DC and macrophages from B-Tg(hLILRB1/hLILRB4) mice.
-
Protein expression in different immune cells

-
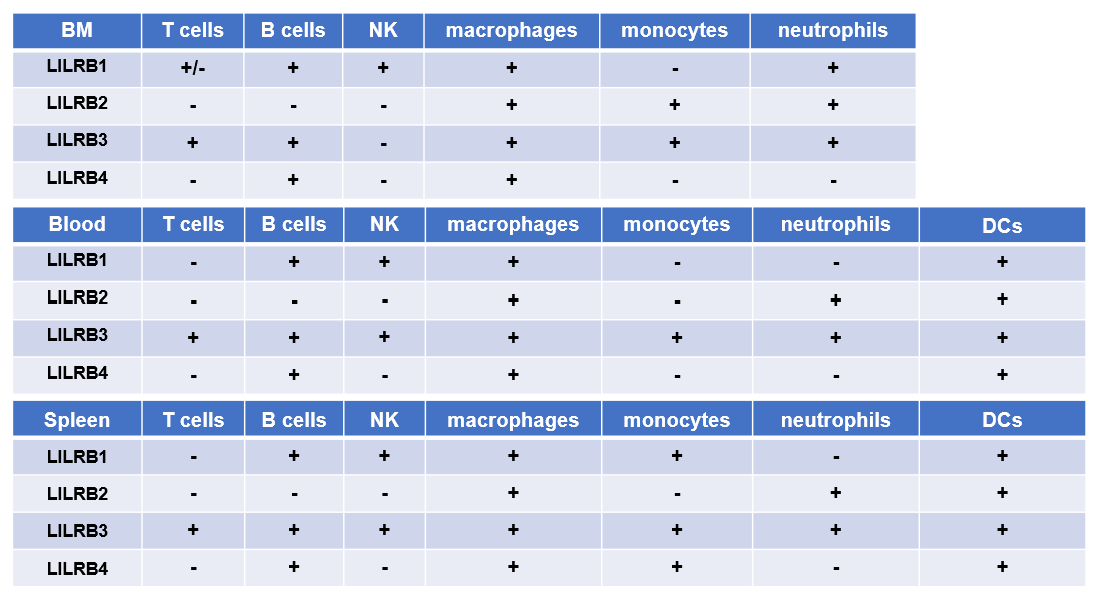
We used flow cytometry to detect the protein expression of various immune cells in bone marrow and spleen. The results are summarized as follows:
-
Analysis of leukocytes cell subpopulation in spleen

-
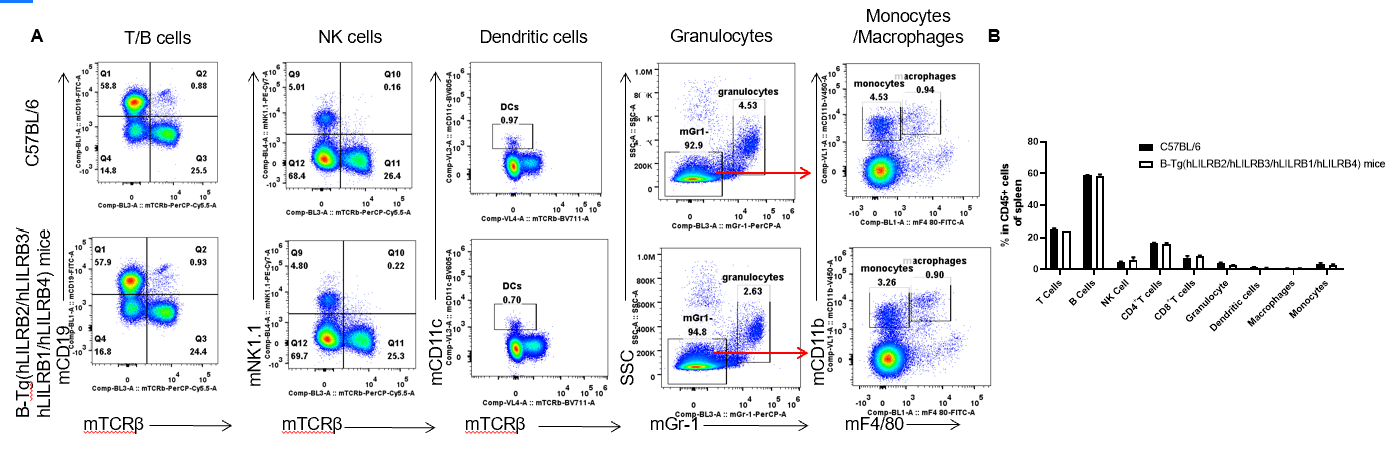
Analysis of spleen leukocyte subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1, LILRB2, LILRB3 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in spleen. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in spleen

-
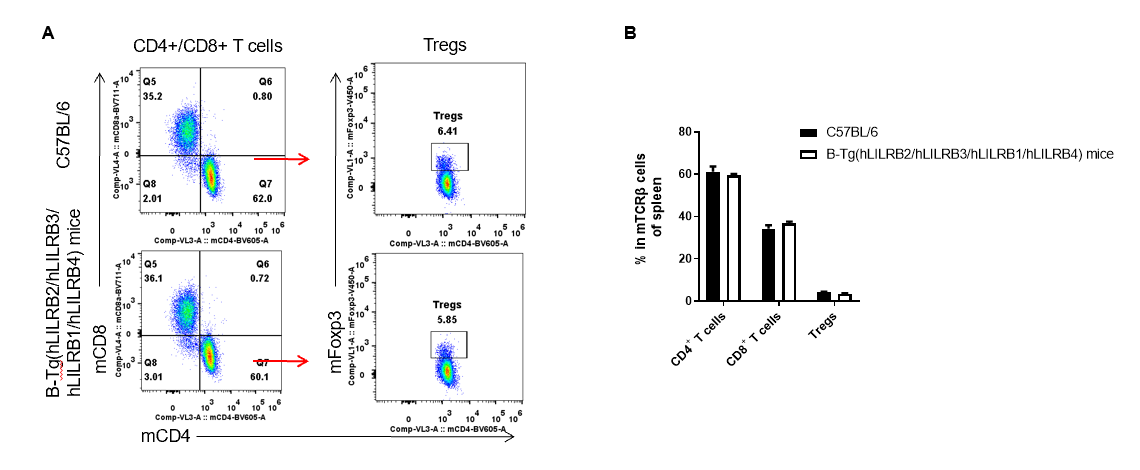
Analysis of spleen T cell subpopulations by FACS. Splenocytes were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the splenocytes was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that introduction of LILRB1, LILRB2, LILRB3 and LILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in spleen. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in LNs

-
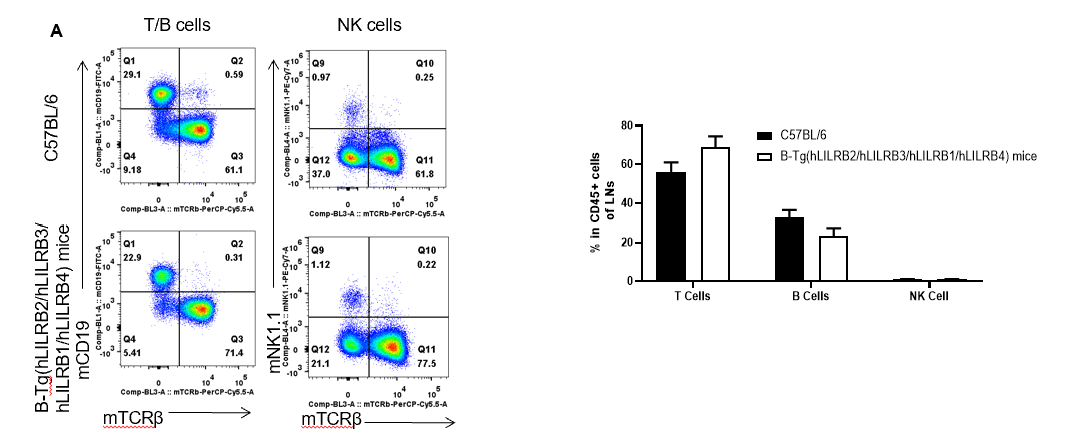
Analysis of thymus leukocyte subpopulations by FACS. LNs (Lymph nodes) were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the LNs was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells and NK cells in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1, LILRB2, LILRB3 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in LNs. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in LNs

-
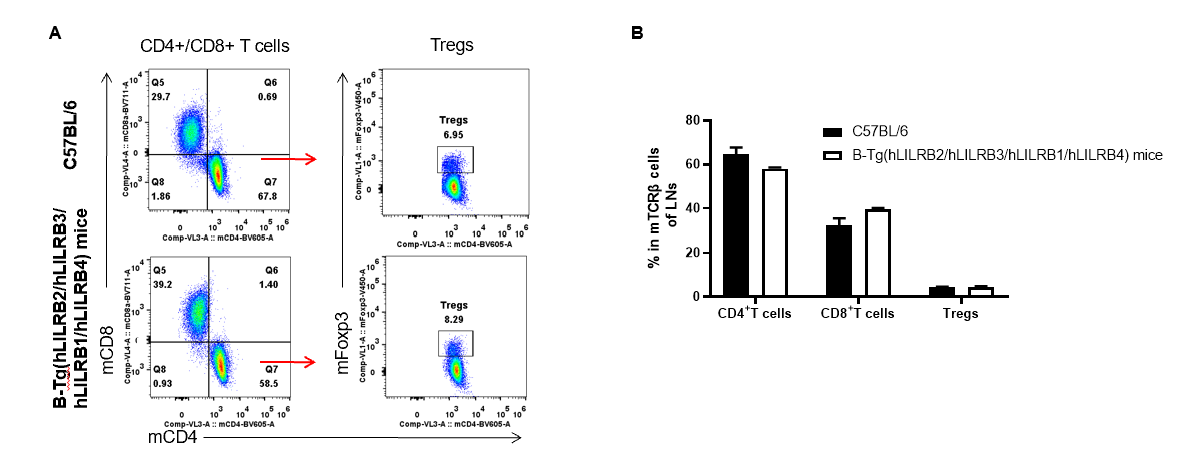
Analysis of thymus T cell subpopulations by FACS. LNs (Lymph nodes) were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the LNs was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that introduction of LILRB1, LILRB2, LILRB3 and LILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in LNs. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in blood

-
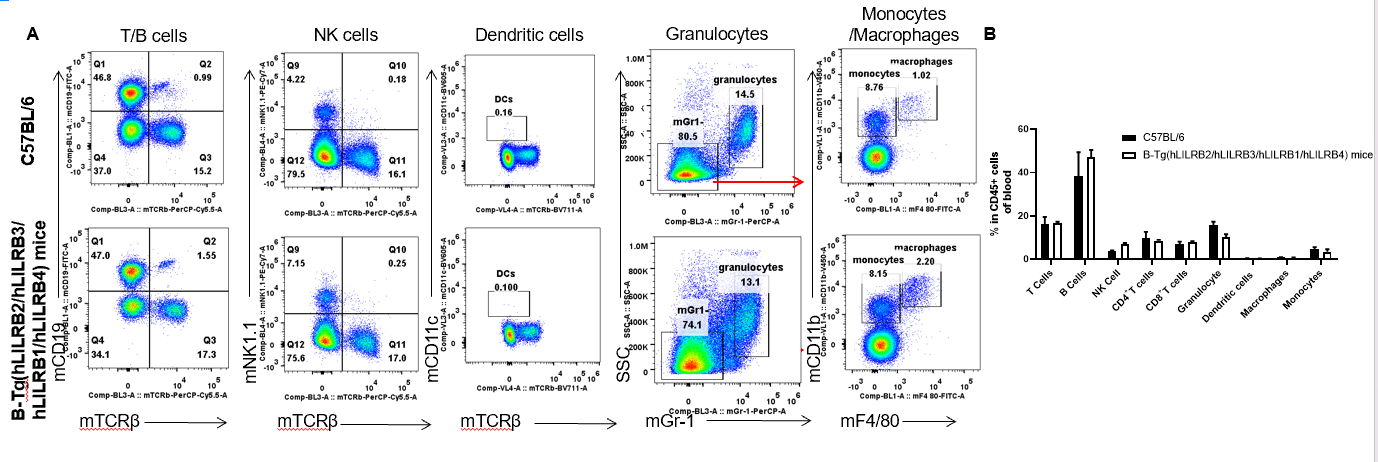
Analysis of blood leukocyte subpopulations by FACS. Blood leukocytes were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the blood leukocytes were performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1, LILRB2, LILRB3 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in blood. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in blood

-
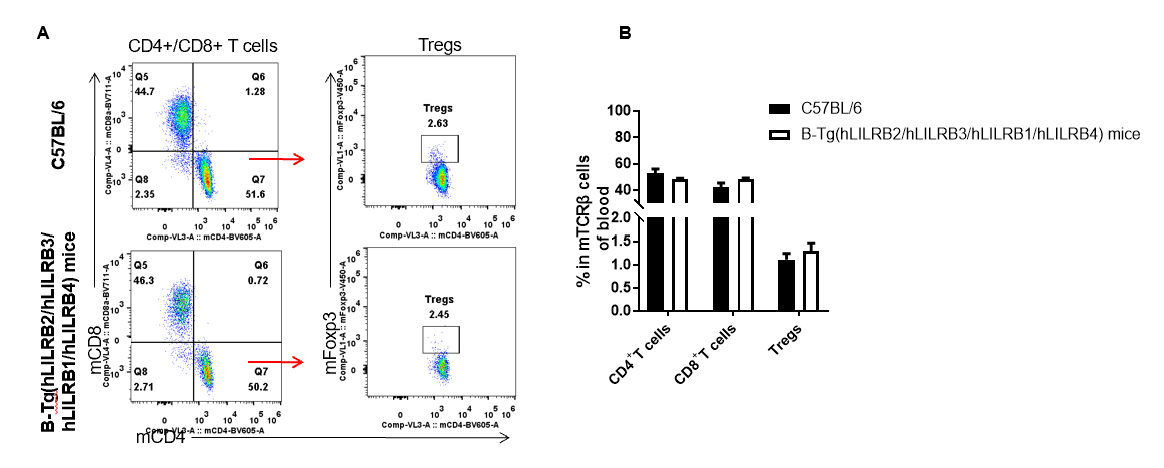
Analysis of blood T cell subpopulations by FACS. Blood leukocytes were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the blood leukocytes were performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that introduction of LILRB1, LILRB2, LILRB3 and LILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in blood. Values are expressed as mean ± SEM.
-
Analysis of leukocytes cell subpopulation in BM

-
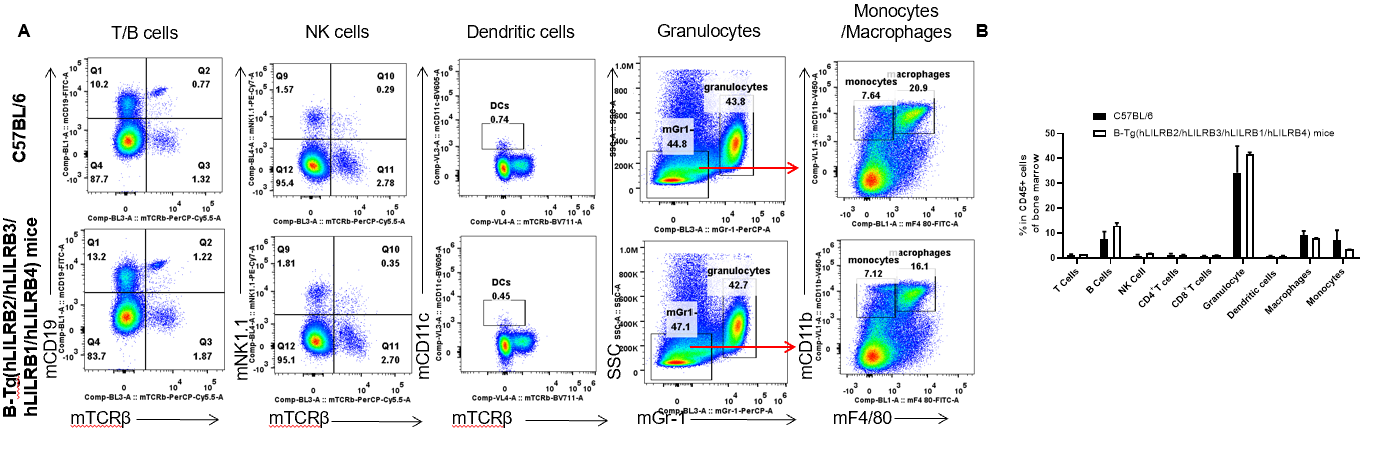
Analysis of bone marrow leukocyte subpopulations by FACS. Bone marrow leukocyte were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the bone marrow leukocyte was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live cells were gated for the CD45+ population and used for further analysis as indicated here. B. Results of FACS analysis. Percent of T cells, B cells, NK cells, dendritic cells, granulocytes, monocytes and macrophages in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that LILRB1, LILRB2, LILRB3 and LILRB4 humanized does not change the overall development, differentiation or distribution of these cell types in bone marrow. Values are expressed as mean ± SEM.
-
Analysis of T cell subpopulation in BM

-
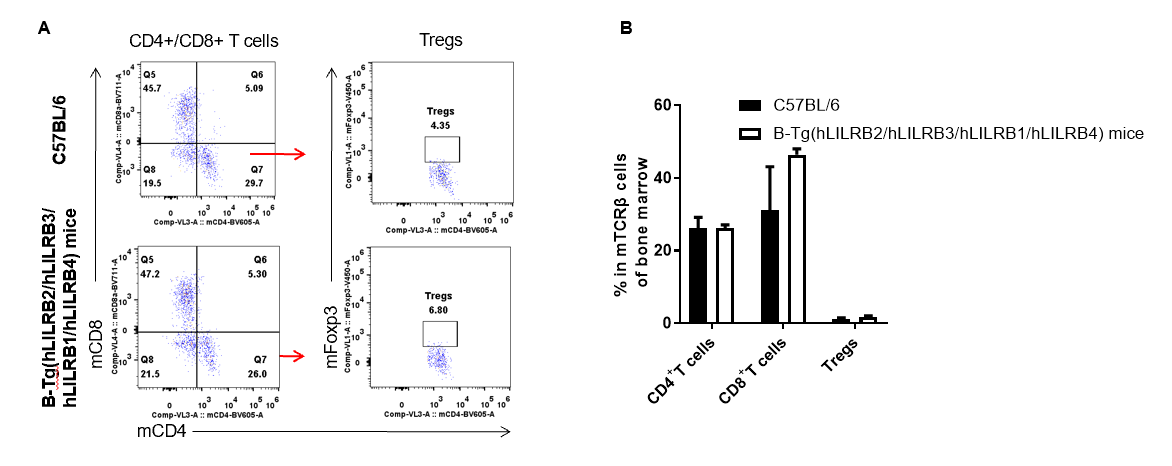
Analysis of bone marrow T cell subpopulations by FACS. Bone marrow leukocyte were isolated from female C57BL/6 and B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice (n=3, 8-week-old). Flow cytometry analysis of the bone marrow leukocyte was performed to assess leukocyte subpopulations. A. Representative FACS plots. Single live CD45+ cells were gated for TCRβ+ T cell population and used for further analysis as indicated here. B. Results of FACS analysis. The percent of CD4+ T cells, CD8+ T cells and Tregs in B-Tg(hLILRB2/hLILRB3/hLILRB1/hLILRB4)mice were similar to those in the C57BL/6 mice, demonstrating that introduction of LILRB1, LILRB2, LILRB3 and LILRB4 does not change the overall development, differentiation or distribution of these T cell subtypes in bone marrow. Values are expressed as mean ± SEM.
-
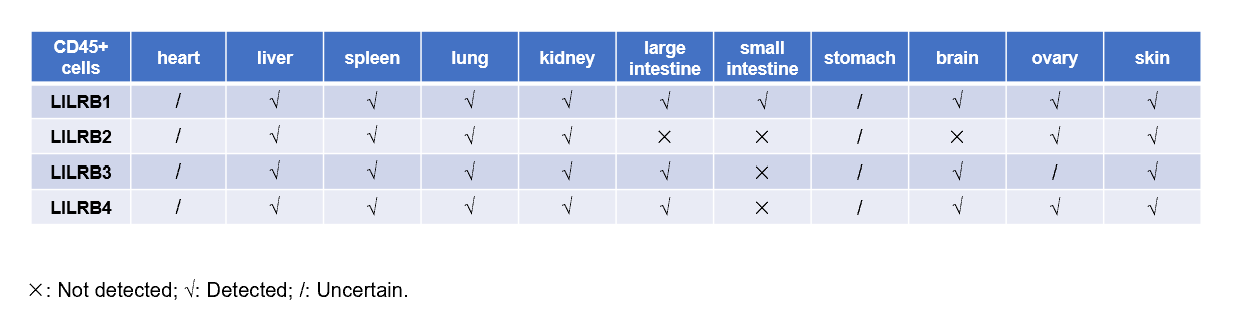
hLILRB1 expression in CD45+ cells

-
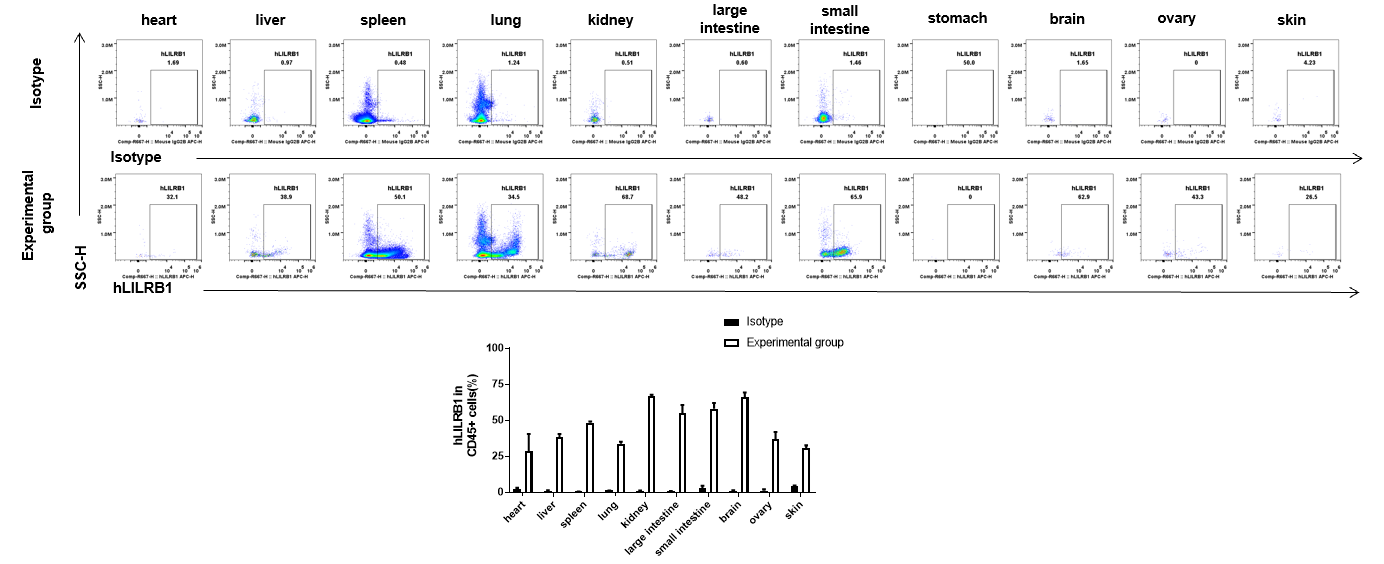
Strain specific hLILRB1 expression analysis in B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice by flow cytometry. Heart, liver, spleen, lung, kidney, large intestine, small intestine, stomach, brain, ovary and skin were collected from B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice, analyzed by flow cytometry with anti-hLILRB1 antibody.
-
hLILRB2 expression in CD45+ cells

-
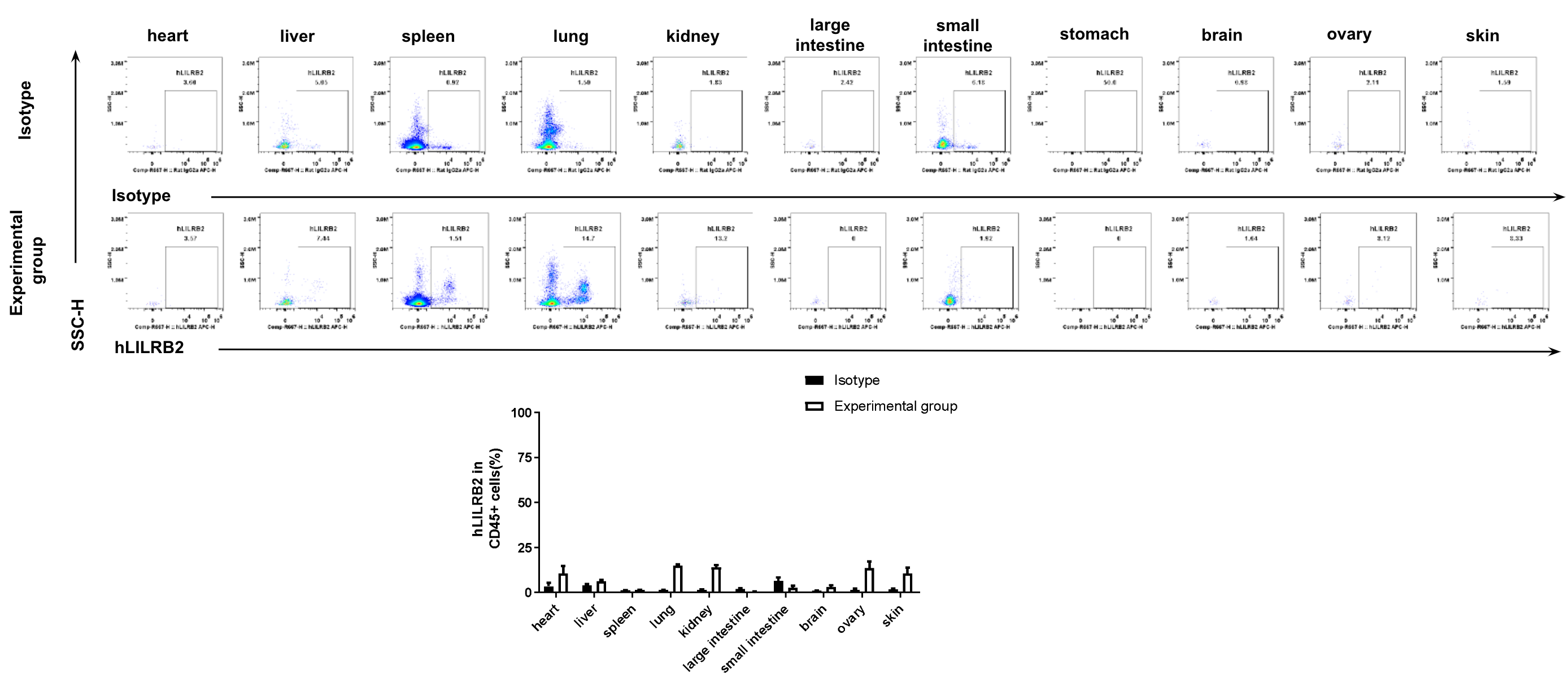
Strain specific hLILRB2 expression analysis in B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice by flow cytometry. Heart, liver, spleen, lung, kidney, large intestine, small intestine, stomach, brain, ovary and skin were collected from B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice, analyzed by flow cytometry with anti-hLILRB2 antibody.
-
hLILRB3 expression in CD45+ cells

-
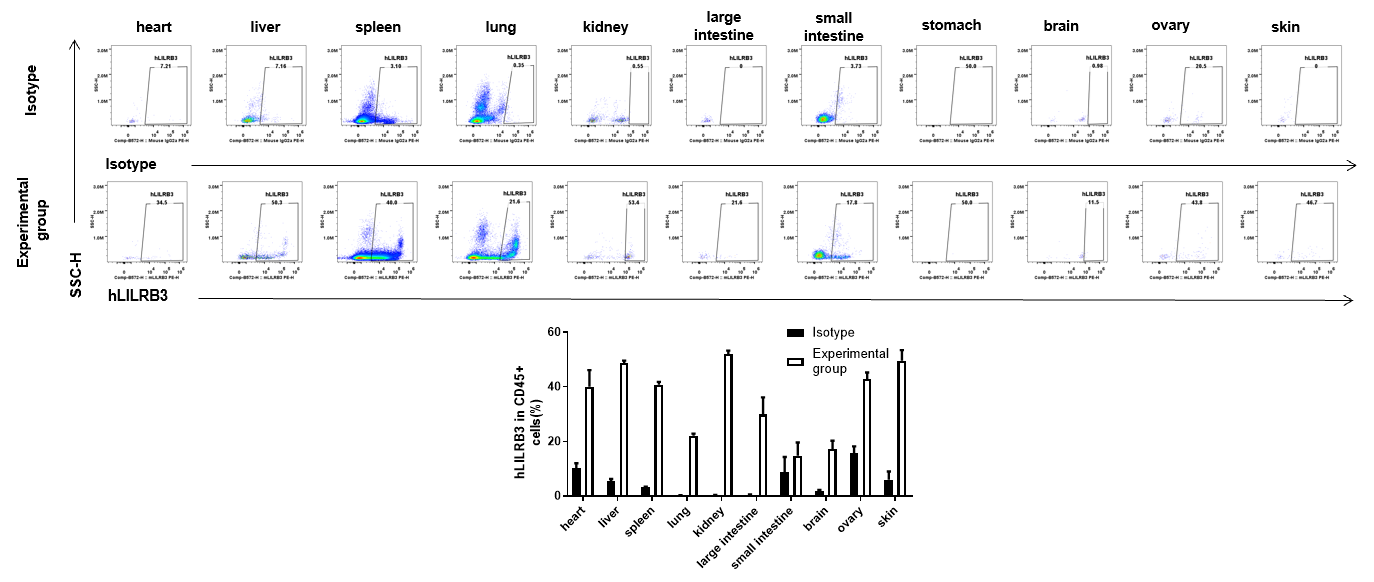
Strain specific hLILRB3 expression analysis in B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice by flow cytometry. Heart, liver, spleen, lung, kidney, large intestine, small intestine, stomach, brain, ovary and skin were collected from B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice, analyzed by flow cytometry with anti-hLILRB3 antibody.
-
hLILRB4 expression in CD45+ cells

-
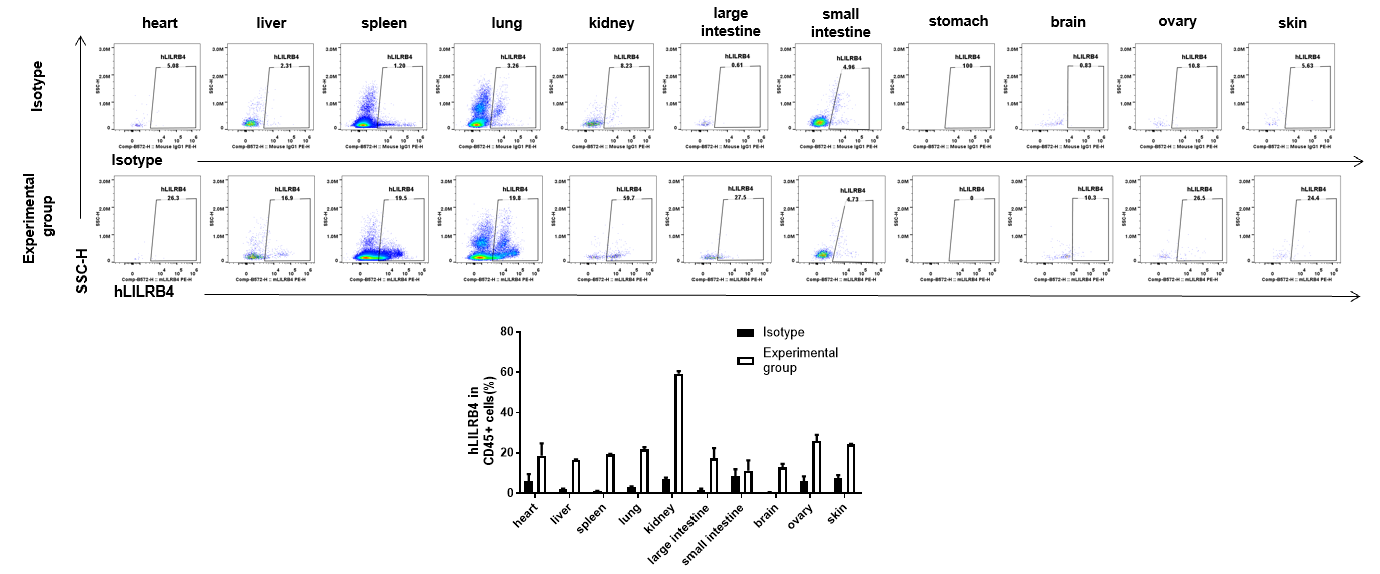
Strain specific hLILRB4 expression analysis in B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice by flow cytometry. Heart, liver, spleen, lung, kidney, large intestine, small intestine, stomach, brain, ovary and skin were collected from B-Tg(hLILRB1/hLILRB2/hLILRB3/hLILRB4) mice, analyzed by flow cytometry with anti-hLILRB4 antibody.
-
Protein expression in different organs

-